BIDS Tutorial Series: HeuDiConv Walkthrough
Introduction
Welcome back to the next tutorial in the tutorial series “Getting Started with BIDS”. In this tutorial, we will be utilizing an off-the-shelf solution called HeuDiConv. This tutorial will illustrate a detailed step-by-step guide on how to use HeuDiConv. The guide will take one subject and iteratively add one session at a time until the dataset is organized and validated. This part will provide separate command line inputs to run HeuDiConv. We will be using DICOMs from the MyConnectome dataset found here and following the BIDS Specification version 1.1.0. Another tutorial has described using HeuDiConv on a different dataset. The next part of this tutorial series will examine another off-the-shelf solution to consider using to convert your dataset into a validated BIDS dataset.
For surfacing questions, we please ask if you may direct them to NeuroStars with the “bids” tag. NeuroStars is an active platform with many BIDS experts.
Table of Contents
- Setting up your environment
- Run HeuDiConv on ses-001 scans to get the dicominfo file
- Examine ses-001 dicominfo file to generate heuristic.py
- Run HeuDiConv on ses-001 scans with heuristic.py and validate
- Run HeuDiConv on ses-005 scans to get the dicominfo file
- Examine ses-005 dicominfo file and update heuristic.py
- Run HeuDiConv on ses-005 scans with updated heuristic.py and validate
- Run HeuDiConv on ses-025 scans to get the dicominfo file
- Examine ses-025 dicominfo file and update heuristic.py
- Run HeuDiConv on ses-025 scans with updated heuristic.py and validate
2A. Guide to using HeuDiConv
Step 1. To begin, we first need to initialize the environment. We will be utilizing Docker for running HeuDiConv. After one has gotten Docker, we can use the command line to download HeuDiConv. The command is shown below. Once that has downloaded, one may begin the conversation
docker pull nipy/heudiconv:latest
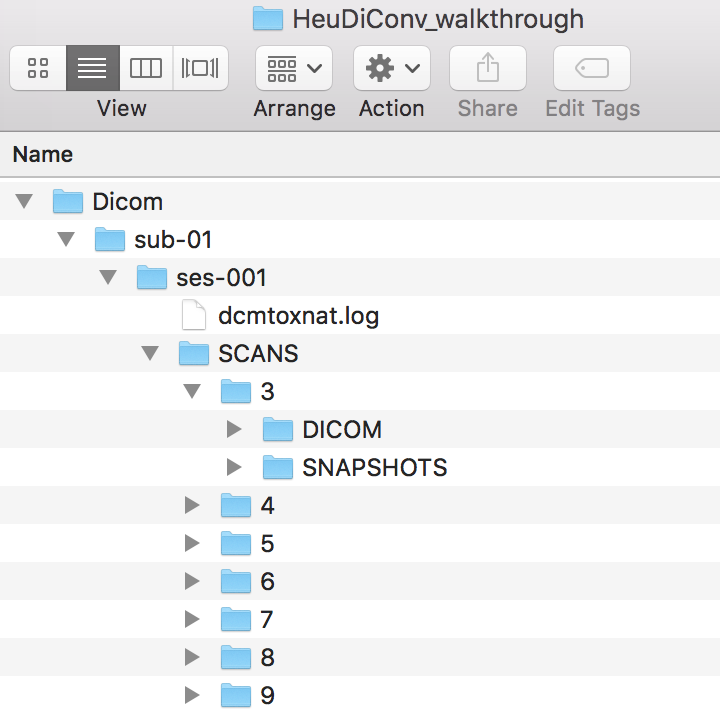
We will be starting with converting ses-001 within sub-01.
Step 2. We are ready to run HeuDiConv. The command syntax can be seen below. Let’s break down this syntax. The ‘docker run –rm -it’ is calling docker. The ‘-v /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough:/base’ is mounting this path and calling it base. This means that base can be used instead of the entire path. The ‘nipy/heudiconv:latest’ is calling the latest version of HeuDiConv. The -d is the dicom directory. Notice we started the path with /base instead of the full path and subject and session are curly bracketed, this is because they are defined later in the call as -s for subject and -ss for session. The -o is where the .heudiconv directory will be, -f is for the heuristic file, and -c is the converter to use.
docker run --rm -it -v /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough:/base nipy/heudiconv:latest -d /base/Dicom/sub-{subject}/ses-{session}/SCANS/*/DICOM/*.dcm -o /base/Nifti/ -f convertall -s 01 -ss 001 -c none --overwrite
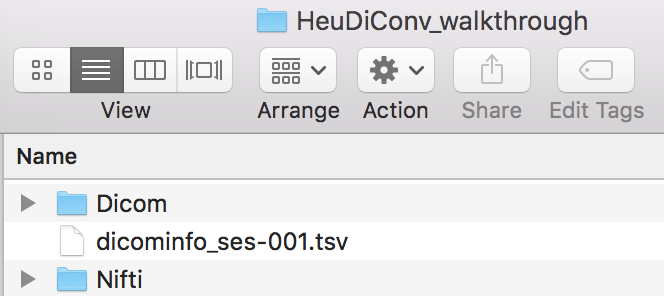
This command will output a hidden .heudiconv folder within the Nifti folder that contains a dicominfo .tsv file used to construct the heuristic file.
Step 3. Now copy the dicominfo .tsv file out of the hidden .heudiconv folder. We temporarily placed this folder at the same level as Dicom and Nifti. This file is used to construct the heuristic.py file.
cp /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough/Nifti/.heudiconv/01/info/dicominfo_ses-001.tsv /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough
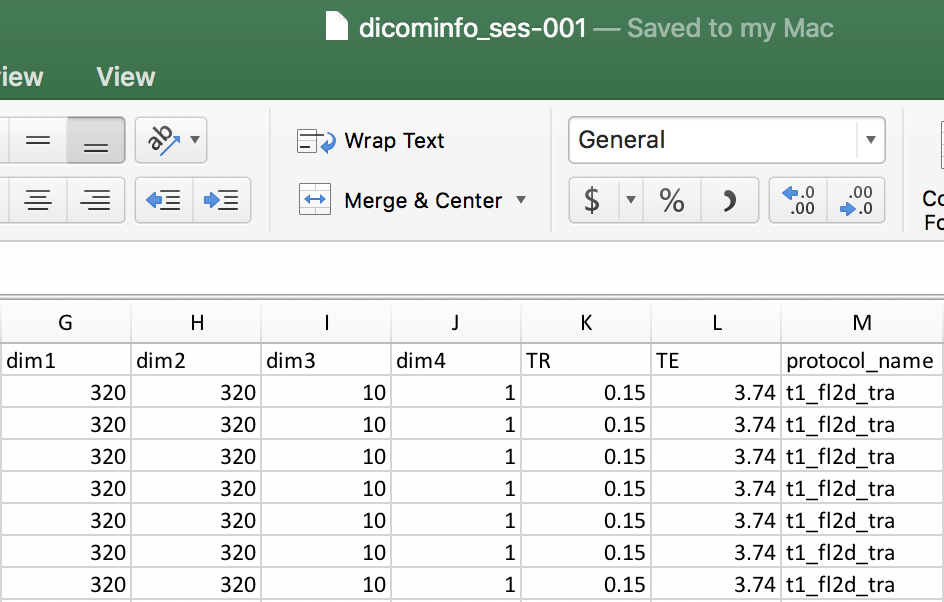
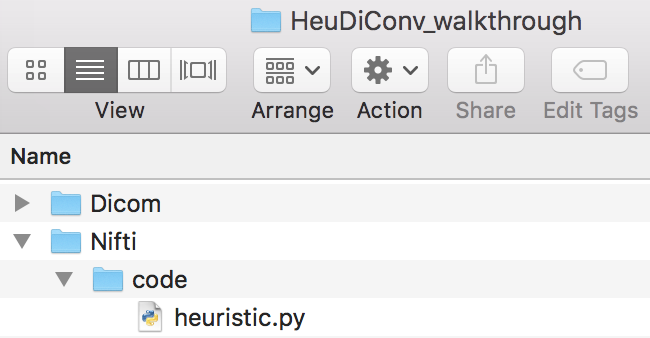
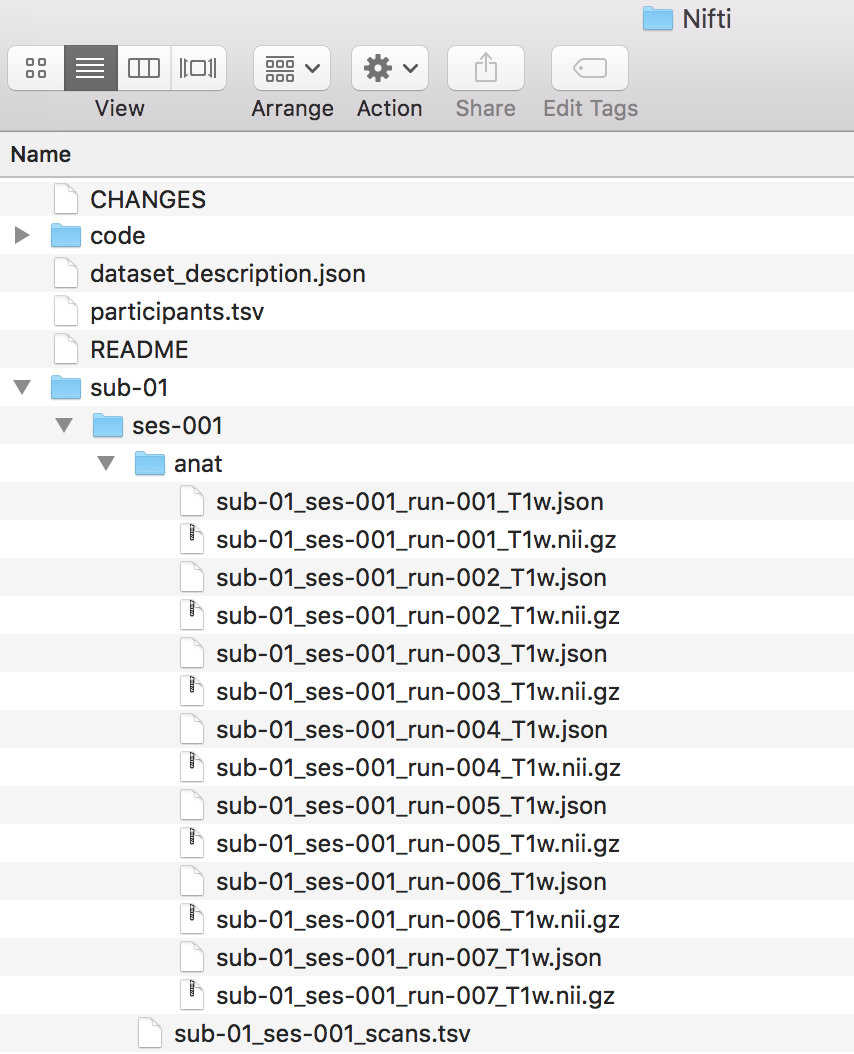
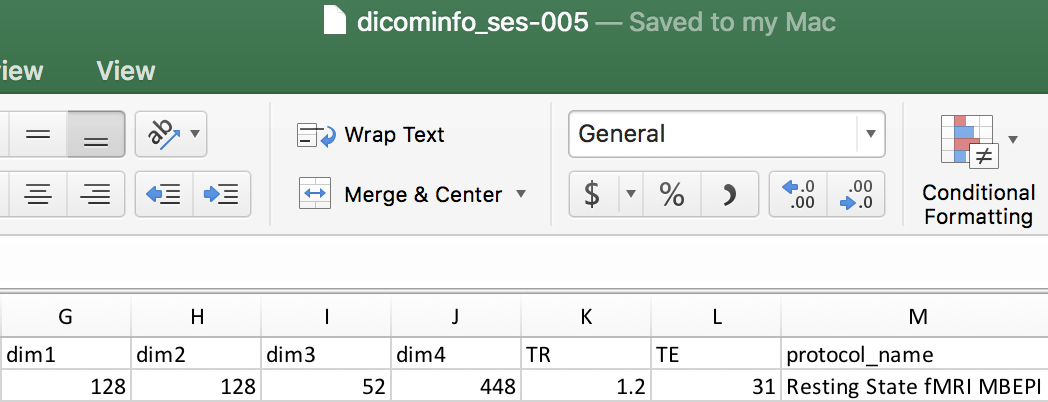
We can open the dicominfo file and evaluate the different columns. Some of the different columns are pictured below. This information will be used to create the heuristic.py file. The heuristic.py file is used to convert and organize the data. Here is an example of how a complete one may look. Our heuristic.py can be seen below. Once the heuristic file has been generated, one may delete the dicominfo .tsv file. Please place the heuristic.py file in a code folder within Nifti, pictured below.
import os
def create_key(template, outtype=('nii.gz',), annotation_classes=None):
if template is None or not template:
raise ValueError('Template must be a valid format string')
return template, outtype, annotation_classes
def infotodict(seqinfo):
"""Heuristic evaluator for determining which runs belong where
allowed template fields - follow python string module:
item: index within category
subject: participant id
seqitem: run number during scanning
subindex: sub index within group
"""
t1w = create_key('sub-{subject}/{session}/anat/sub-{subject}_{session}_run-00{item:01d}_T1w')
info = {t1w: []}
for idx, s in enumerate(seqinfo):
if (s.dim1 == 320) and (s.dim2 == 320) and ('t1_fl2d_tra' in s.protocol_name):
info[t1w].append(s.series_id)
return info
Step 4. Now that we have the heuristic.py script we can run HeuDiConv again, but changing -c from none to dcm2niix and adding -b. The call is seen below.
docker run --rm -it -v /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough:/base nipy/heudiconv:latest -d /base/Dicom/sub-{subject}/ses-{session}/SCANS/*/DICOM/*.dcm -o /base/Nifti/ -f /base/Nifti/code/heuristic.py -s 01 -ss 001 -c dcm2niix -b --overwrite
Once HeuDiConv completes running, the output is pictured below. We are ready to validate this dataset.
We find this is a valid BIDS dataset.
Step 5. We will now be adding in ses-005.
Similar to the approach taken for converting and organizing the ses-001 images, we will be running HeuDiConv to get the dicominfo .tsv file. To do this, the syntax will follow closely to the call used for getting the ses-001 dicominfo file.
docker run --rm -it -v /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough:/base nipy/heudiconv:latest -d /base/Dicom/sub-{subject}/ses-{session}/SCANS/*/DICOM/*.dcm -o /base/Nifti/ -f convertall -s 01 -ss 005 -c none --overwrite
This will output the dicominfo .tsv file for ses-005. Please copy the dicominfo .tsv file out of the .heudiconv directory.
Step 6. We can begin adding to the heuristic.py file to convert and organize the diffusion scans. To do this, we look at the dicominfo.tsv file and determine the condition to organize the scan. This will be added to the heuristic.py file.
import os
def create_key(template, outtype=('nii.gz',), annotation_classes=None):
if template is None or not template:
raise ValueError('Template must be a valid format string')
return template, outtype, annotation_classes
def infotodict(seqinfo):
"""Heuristic evaluator for determining which runs belong where
allowed template fields - follow python string module:
item: index within category
subject: participant id
seqitem: run number during scanning
subindex: sub index within group
"""
t1w = create_key('sub-{subject}/{session}/anat/sub-{subject}_{session}_run-00{item:01d}_T1w')
func_rest = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-00{item:01d}_bold')
info = {t1w: [], func_rest: []}
for idx, s in enumerate(seqinfo):
if (s.dim1 == 320) and (s.dim2 == 320) and ('t1_fl2d_tra' in s.protocol_name):
info[t1w].append(s.series_id)
if (s.dim1 == 128) and (s.dim2 == 128) and ('Resting State fMRI MBEPI' in s.protocol_name):
info[func_rest].append(s.series_id)
return info
Step 7. Now that we have the updated heuristic.py file, we can delete the dicominfo.tsv file and run HeuDiConv again to convert and organize ses-005.
docker run --rm -it -v /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough:/base nipy/heudiconv:latest -d /base/Dicom/sub-{subject}/ses-{session}/SCANS/*/DICOM/*.dcm -o /base/Nifti/ -f /base/Nifti/code/heuristic.py -s 01 -ss 005 -c dcm2niix -b --overwrite
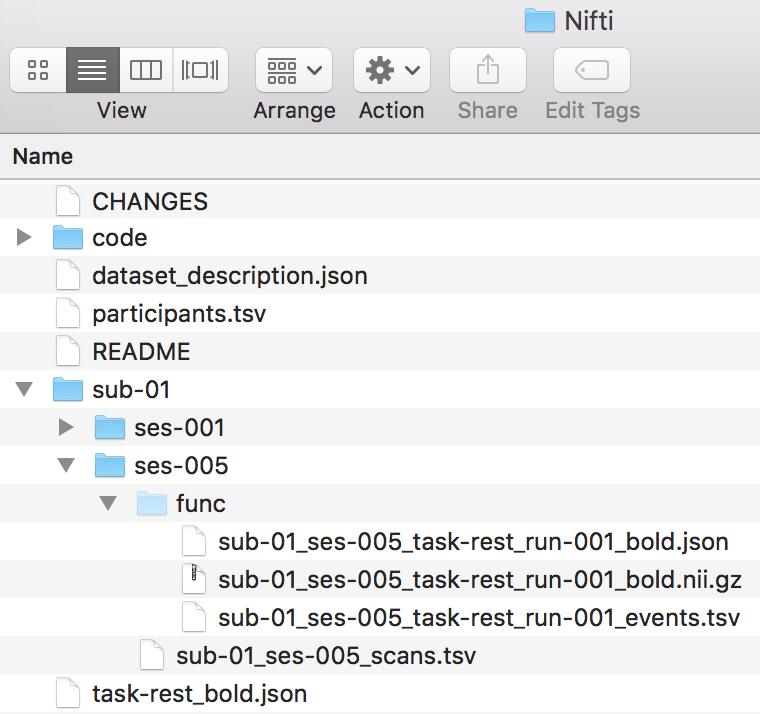
We find that HeuDiConv converted and organized the scan, so we are ready to validate our dataset. In addition, shown below is the file structure up to this point.
We find this is a valid BIDS dataset.
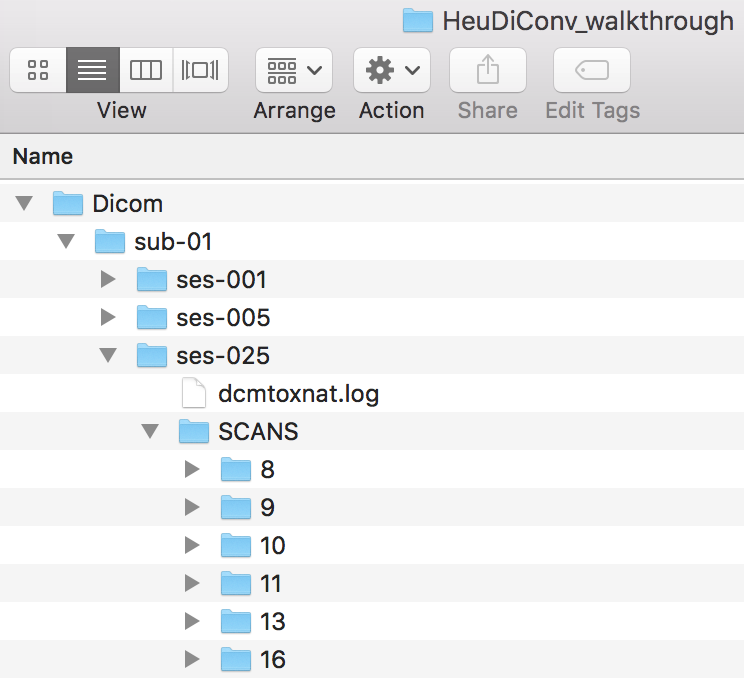
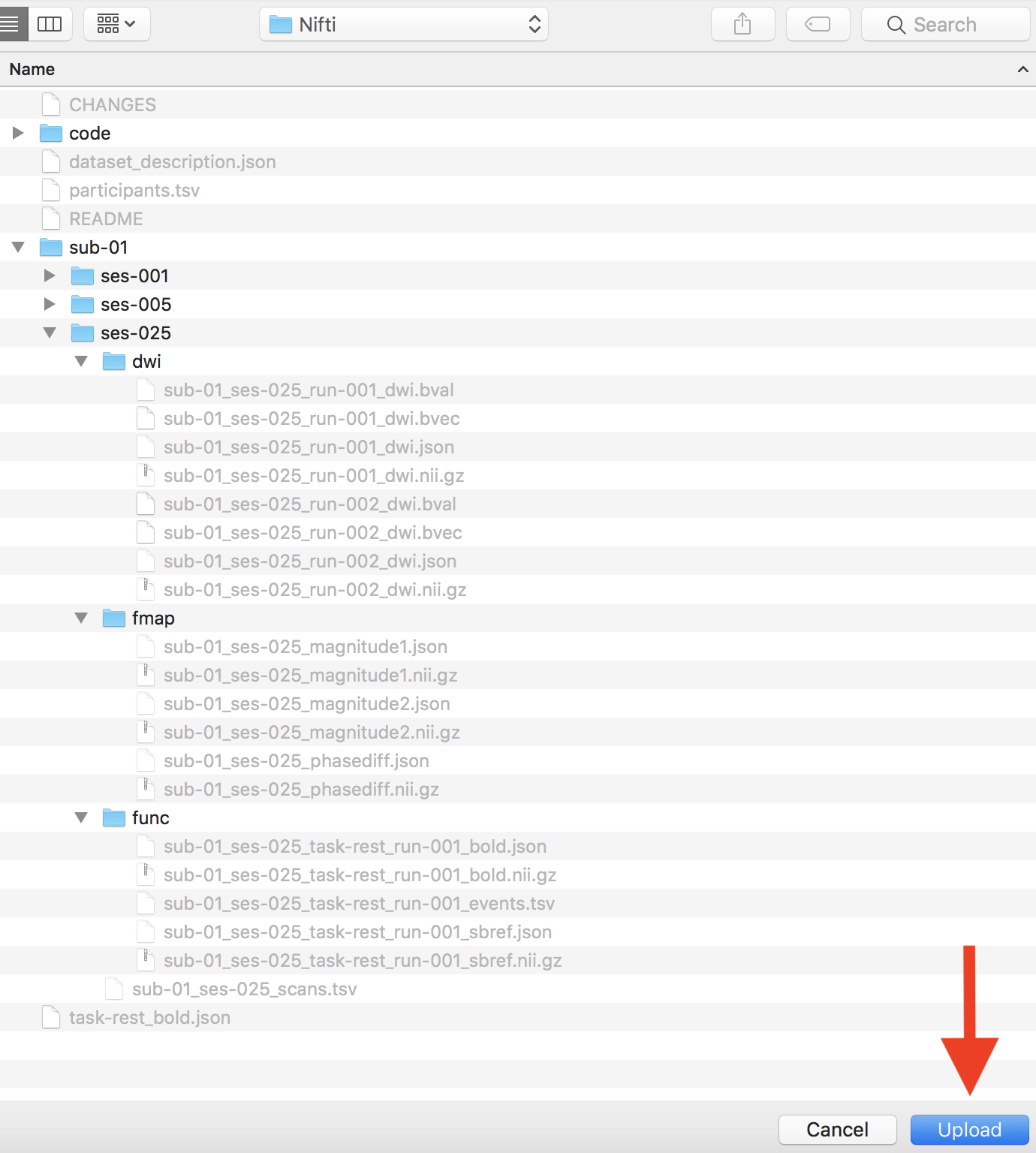
Step 8. Now we are ready to add in the ses-025 scans. The Dicom structure is pictured below.
Following the same approach taken for ses-001 and ses-005 conversation and organization, we will run HeuDiConv to get the dicominfo .tsv file. To do this, the command will follow closely to the HeuDiConv call used above.
This will run and output the dicominfo .tsv file. Copy the file out of the .heudiconv directory.
docker run --rm -it -v /Users/franklinfeingold/Desktop/HeuDiConv_walkthrough:/base nipy/heudiconv:latest -d /base/Dicom/sub-{subject}/ses-{session}/SCANS/*/DICOM/*.dcm -o /base/Nifti/ -f convertall -s 01 -ss 025 -c none --overwrite
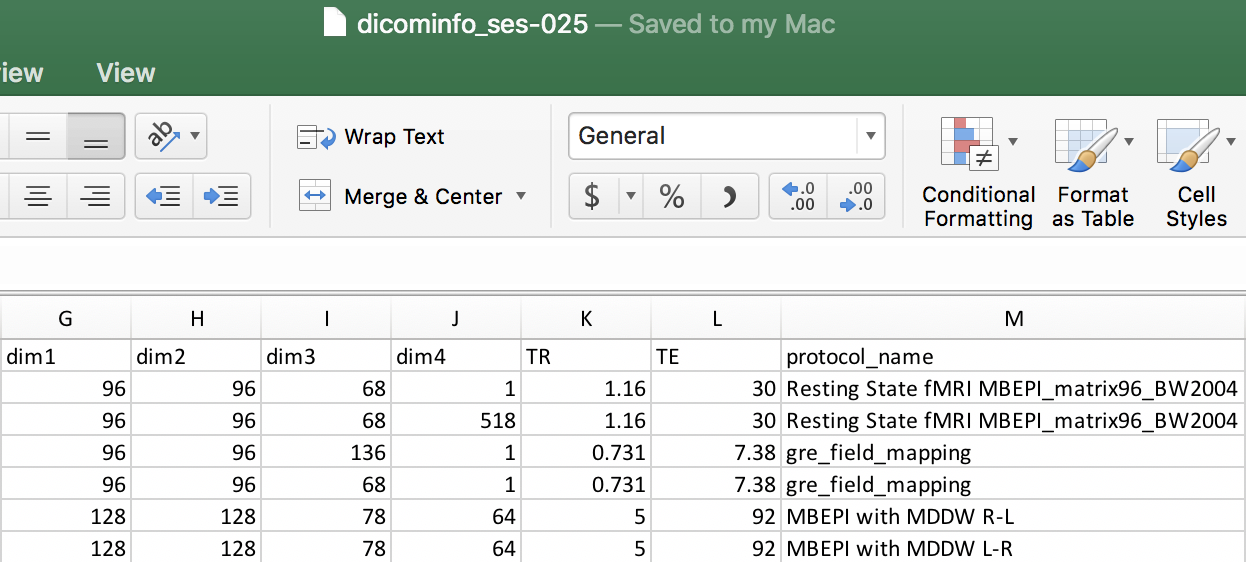
Step 9. We can determine the organizational rules for each of the scans from the dicominfo file and add them to the heuristic.py file. This follows the same process done for determining the rules to organize the ses-001 and ses-005 scans. We have pictured the dicominfo file and the completed heuristic.py file.
import os
def create_key(template, outtype=('nii.gz',), annotation_classes=None):
if template is None or not template:
raise ValueError('Template must be a valid format string')
return template, outtype, annotation_classes
def infotodict(seqinfo):
"""Heuristic evaluator for determining which runs belong where
allowed template fields - follow python string module:
item: index within category
subject: participant id
seqitem: run number during scanning
subindex: sub index within group
"""
t1w = create_key('sub-{subject}/{session}/anat/sub-{subject}_{session}_run-00{item:01d}_T1w')
func_rest = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-00{item:01d}_bold')
func_rest_matrix96 = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-00{item:01d}_bold')
func_rest_matrix96_sbref = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-00{item:01d}_sbref')
fmap_mag = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_magnitude')
fmap_phase = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_phasediff')
dwi = create_key('sub-{subject}/{session}/dwi/sub-{subject}_{session}_run-00{item:01d}_dwi')
info = {t1w: [], func_rest: [], func_rest_matrix96: [], func_rest_matrix96_sbref: [], fmap_mag: [], fmap_phase: [], dwi: []}
for idx, s in enumerate(seqinfo):
if (s.dim1 == 320) and (s.dim2 == 320) and ('t1_fl2d_tra' in s.protocol_name):
info[t1w].append(s.series_id)
if (s.dim1 == 128) and (s.dim2 == 128) and ('Resting State fMRI MBEPI' in s.protocol_name):
info[func_rest].append(s.series_id)
if (s.dim1 == 96) and (s.dim4 == 518) and ('Resting State fMRI MBEPI_matrix96_BW2004' in s.protocol_name):
info[func_rest_matrix96].append(s.series_id)
if (s.dim1 == 96) and (s.dim4 == 1) and ('Resting State fMRI MBEPI_matrix96_BW2004' in s.protocol_name):
info[func_rest_matrix96_sbref].append(s.series_id)
if (s.dim3 == 136) and (s.dim4 == 1) and ('gre_field_mapping' in s.protocol_name):
info[fmap_mag] = [s.series_id]
if (s.dim3 == 68) and (s.dim4 == 1) and ('gre_field_mapping' in s.protocol_name):
info[fmap_phase] = [s.series_id]
if (s.dim2 == 128) and (s.dim4 == 64):
info[dwi].append(s.series_id)
return info
Step 10. We have updated the heuristic.py file, so we can delete the dicominfo .tsv file and run HeuDiConv again to convert and organize ses-025. The command to do this is seen below.
docker run --rm -it -v /Users/franklinfeingold/Desktop/Heudiconv_walkthrough:/base nipy/heudiconv:latest -d /base/Dicom/sub-{subject}/ses-{session}/SCANS/*/DICOM/*.dcm -o /base/Nifti/ -f /base/Nifti/code/heuristic.py -s 01 -ss 025 -c dcm2niix -b --overwrite
After running HeuDiConv for ses-025, we are ready to validate our dataset. We have pictured below the ses-025 Nifti file structure.
We find this is a valid BIDS dataset!
This tutorial demonstrated how to utilize HeuDiConv for converting DICOMs into a validated BIDS dataset. In the next part, we will walk through another off-the-shelf solution to consider for converting your datasets.



Is there a tutorial for using HeuDiConv without Docker? Would be extremely helpful! Thanks in advance.
Hi Dylan,
I am not aware of a tutorial that doesn’t use Docker. On the Heudiconv github (https://github.com/nipy/heudiconv) they describe how to set it up locally. From there the tutorial should be valid, but instead of running the container you are calling Heudiconv directly. The steps should be the same.
I hope that helps!
Franklin
Hi,
thanks for the great tutorial. It has been very helpful. A minor issue, but heudiconv generates a file called convertall.py instead of heuristic.py (step 3).
Hi Dianne,
Great to hear this tutorial has been helpful! Thank you for bring up this issue. Heudiconv will generate the convertall.py within the heudiconv hidden folder on the first pass. This is because it is the initial config file you are running to generate the initial dicominfo tsv file to help create your heuristic.py file. The heuristic.py file is what we are generating so heudiconv can organize the imaging files according to the rules defined in the heuristic file. When one runs it the second time, the heuristic.py file will be generated in place of the convertall.py file.
I hope this clarifies this issue,
Franklin
Thank you for putting all of these together. They have been so helpful! Is anyone aware of a tutorial using singularity, rather than docker, with heudiconv?
Hi Franklin,
I appreciate your response. I have modified and run convertall.py many times now (it works well, thanks to this useful tutorial). I have never seen this mysterious heuristic.py…not in the hidden .heudiconv directory, not in any subdirectories of Code…perhaps it is created temporarily in the docker file and then gone when the instance finishes running? I am mystified…
I am actually wondering now if there is any information on running reproin with docker…it seems to be a bit different, and I’m struggling to guess how to make it work. Any suggestions?
Thanks for your time.
Maybe hueristic.py is just the name you chose? I just copied convertall.py to Code…it helped me to modify the template.
WRT reproin, never mind…this is working ; )
docker run –rm -it -v ${PWD}:/base nipy/heudiconv:latest -f reproin –bids -o /base/Nifti –files /base/Dicom/phantom-1
Hi Lauren,
Great to hear the tutorials have been helpful! I am not aware of any, but asking over on their github (https://github.com/nipy/heudiconv), they may be able to point you in a good direction to using heudiconv with singularity.
Thank you,
Franklin
Hi Dianne,
Thank you for your follow up! I apologize for the confusion! The heuristic.py file is generated by you, heudiconv will not generate this. Heudiconv generates the dicominfo.tsv file for you to use to generate the rules for the heuristic.py file. For example in step 3 where I have shown my dicominfo file and under it what my heuristic.py file (that I created) would look like. I’m happy to hear that reproin worked!
I hope this helps clear up the confusion!
Franklin
Hi there,
I am trying to use the BIDS validator, but I always get an error that says:
1: [ERR] You have to define ‘TaskName’ for this file. (code: 50 – TASK_NAME_MUST_DEFINE)
./sub-01/ses-005/func/sub-01_ses-005_task-rest_run-001_bold.nii.gz
./sub-01/ses-025/func/sub-01_ses-025_task-rest_run-001_bold.nii.gz
Has anyone experienced this behavior? How can I fix it?
Thanks for any help on this.
Hi Hugo,
Thank you for your question. This error pertains to defining the TaskName in your json file associated with those files. For example this would be the ./sub-01/ses-005/func/sub-01_ses-005_task-rest_run-001_bold.json file. More information regarding adding the TaskName can be found here: http://reproducibility.stanford.edu/bids-tutorial-series-part-1a/#man14 . It is the highlighted portion adding the TaskName to the json file.
Thank you,
Franklin
Hi –
Very basic question: step 1 in this tutorial yields the following error for me:
docker pull nipy/heudiconv:latest
Error response from daemon: manifest for nipy/heudiconv:latest not found
I tried installing HeuDiConv with pip, which worked fine, and then tried to avoiding the :latest tag in other steps, but it looks like it’s automatically built in even when not explicitly included, so it kept giving the same error. Any advice?
Hi Brian,
Thank you for your question. By chance was docker running when you received this error? I have found this error is typically associated with docker not running with trying to use docker. The pip error is interesting – it could be worth raising it on their github: https://github.com/nipy/heudiconv.
Thank you,
Franklin
@Brian Odegaard I encountered into the same error and used the following command to make it work `docker pull nipy/heudiconv:debian`
Thank you for this great tutorial. I am following the steps on the example dataset, however I can not create .tsv file. when i run step 2, i get this –
INFO: Running heudiconv version 0.6.0.dev1
INFO: Need to process 0 study sessions
and there is nothing in output folder.
Thank you for your question! This appears to be an issue with the mounting of directories and HeuDiConv not reading them. This is seen from the message `INFO: Need to process 0 study sessions`. Confirming the paths to the files and the subject and session keys.
Hi,
Thank you for this tutorial.
I am getting an error that I am struggling to understand. Yesterday, it worked perfectly until step 4.
However, today it’s more difficult because I have an issue “INFO: Need to process 0 study sessions”.
I already had this issue at the beginning but I did not really understand how I solved it .
Did I miss an important step before applying step 2 ? Something about re-initialisation or something similar ?
Best,
Simon
It ‘s has been solved after rebooting my computer but I did not understand the reason …
Hi Simon,
Good to hear this was resolved! This case may be something the HeuDiConv team could provide guidance on.
Thank you,
Franklin
Thank you for the great tutorial.
If I want to validate session 5 I get the following error:
JSON_SCHEMA_VALIDATION_ERROR
sub-01_ses-005_task-rest_run-001_bold.json43.582 KB
Location:
Nifti/sub-01/ses-005/func/sub-01_ses-005_task-rest_run-001_bold.json
Reason:
Invalid JSON file. The file is not formatted according the schema.
Evidence:
.CogAtlasID should match format “uri”
task-rest_bold.json43.631 KB
Location:
Nifti/task-rest_bold.json
Reason:
Invalid JSON file. The file is not formatted according the schema.
Evidence:
.CogAtlasID should match format “uri”
I am not sure how to solve that issue?
Thank you again for your help.
Carina
Hi Carina,
Thank you for your message and I apologize for my very delayed reply. If you may please direct questions over to NeuroStars (https://neurostars.org/tag/bids) with the “bids” tag. That is a more active platform for supporting the community to adopt BIDS.
Thank you,
Franklin
https://suggestrank.com is doing a great car comparison. check it out once.
size expert online size expert online A time bar lasting round 15 in-recreation minutes
will seem excessive of the pot, and the participant should add each different ingredient to the Set earlier than the time runs out.
The participant also can end cooking earlier than the timer runs out by interacting with the Set.
The cooking time will differ primarily based on amount.
Recipe substances are misplaced on a failed recipe try, and can end in Food Waste.
Cooking Set can mix varied cooking substances into new consumables.
You can also make an enormous cooking mistake if you do not try this.
Ensure that the components you’re shopping for don’t comprise excessive quantities of sugar or
sodium, as these each may cause well being issues if overdone.
Add the mince beef, beef inventory cube (crumbled), salt and black pepper stir all of the components till
the beef is barely brown and put aside. However, a few years in the past in England, in home science class, we have been taught to cook shepherds pie utilizing minced beef.