BIDS Tutorial Series: Automate the Introductory Walkthrough
Introduction
Welcome to part 1B of the tutorial series “Getting Started with BIDS”. This tutorial will illustrate how one can automate the conversation of DICOMs into a valid BIDS dataset. This tutorial will follow the same workflow detailed in part 1A of this tutorial series. We will be using DICOMs from the Nathan Kline Institute (NKI) Rockland Sample – Multiband Imaging Test-Retest Pilot Dataset. We will be following the specifications described in the BIDS Specification version 1.0.2. If you are running into issues, please post your questions on NeuroStars with the bids tag. The next parts of this tutorial series will examine off-the-shelf solutions to consider using to convert your dataset into the BIDS standard.
Table of Contents
B. Automated custom conversation
- Initialize script and create the dataset_description file
- Create anatomical folders and convert dicoms
- Rename anatomical files
- Organize anatomical files and validate
- Create diffusion folders and convert dicoms
- Rename diffusion files
- Organize diffusion files and validate
- Create functional folders and convert dicoms
- Rename and organize functional files
- Task event tsv files
- Fix errors
- Validate and add participant
The automated custom solution is going through the same process done in part 1A, but with a script. The automated custom solution is written in a bash shell script. This script is dependent upon homebrew, jo, jq, dcm2niix (jo and jq can be installed from homebrew). The script assumes that the Dicom folder with subjects needed to be converted exists. Each code snippet is part of the larger script.
Step 1. To begin, we need to define our paths and create the Nifti directory. These paths need to be changed according to what your paths are. These path variables are important to remember because these path variables are called throughout the script. (i.e. the variable niidir is defined as the path: /Users/franklinfeingold/Desktop/NKI_script/Nifti)
#!/bin/bash set -e ####Defining pathways toplvl=/Users/franklinfeingold/Desktop/NKI_script dcmdir=/Users/franklinfeingold/Desktop/NKI_script/Dicom dcm2niidir=/Users/franklinfeingold/Desktop/dcm2niix_3-Jan-2018_mac #Create nifti directory mkdir ${toplvl}/Nifti niidir=${toplvl}/Nifti
Then we can generate the dataset_description.json file.
###Create dataset_description.json
jo -p "Name"="NKI-Rockland Sample - Multiband Imaging Test-Retest Pilot Dataset" "BIDSVersion"="1.0.2" >> ${niidir}/dataset_description.json
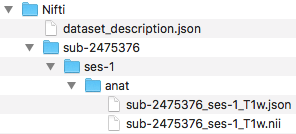
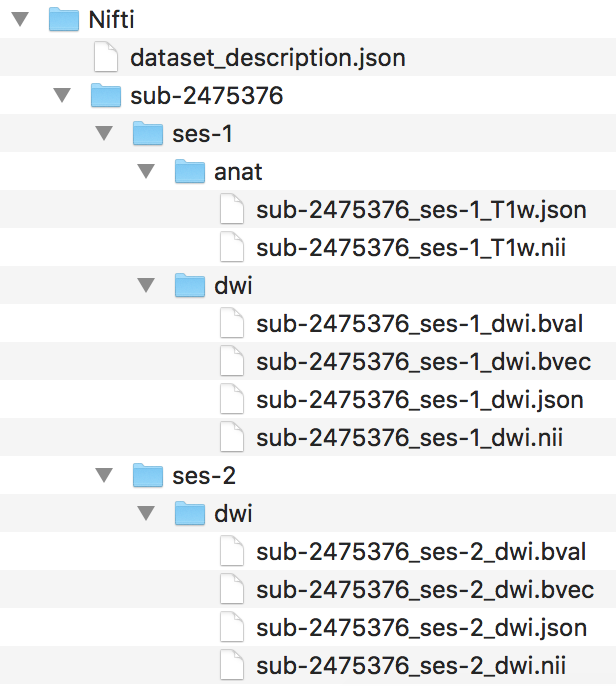
Step 2. To begin the workflow, the loop prints out the subject currently being processed and creates the session and anat folder within the subject folder. Then we convert with dcm2niix the anatomical dicoms into the nifti and json file, and output them in the subject folder within the Nifti folder (output pictured below). Please note that the converter input is the anat folder within the subjects Dicom folder.
The for loop can iterate through every subject defined in the Dicom directory. For this walkthrough, we are only running through participant 2475376. One may add participant 3893245 when comfortable with the workflow. A participant will go through the entire workflow before the next one begins.
####Anatomical Organization#### for subj in 2475376; do echo "Processing subject $subj" ###Create structure mkdir -p ${niidir}/sub-${subj}/ses-1/anat ###Convert dcm to nii #Only convert the Dicom folder anat for direcs in anat; do ${dcm2niidir}/dcm2niix -o ${niidir}/sub-${subj} -f ${subj}_%f_%p ${dcmdir}/${subj}/${direcs} done
We changed the directory to where the converted nifti images are. In that directory, we can more easily change the filenames for the anat nii and json files.
#Changing directory into the subject folder
cd ${niidir}/sub-${subj}
Step 3. Now we can rename the anat files, following the same rule applied in step 6 of part A. This code snippet will capture the number of anat files needed to be changed, go through each anat file and rename it to the valid BIDS filename.
###Change filenames ##Rename anat files #Example filename: 2475376_anat_MPRAGE #BIDS filename: sub-2475376_ses-1_T1w #Capture the number of anat files to change anatfiles=$(ls -1 *MPRAGE* | wc -l) for ((i=1;i<=${anatfiles};i++)); do Anat=$(ls *MPRAGE*) #This is to refresh the Anat variable, if this is not in the loop, each iteration a new "No such file or directory error", this is because the filename was changed. tempanat=$(ls -1 $Anat | sed '1q;d') #Capture new file to change tempanatext="${tempanat##*.}" tempanatfile="${tempanat%.*}" mv ${tempanatfile}.${tempanatext} sub-${subj}_ses-1_T1w.${tempanatext} echo "${tempanat} changed to sub-${subj}_ses-1_T1w.${tempanatext}" done
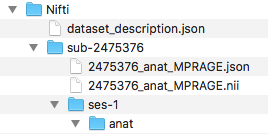
Step 4. Next, we can organize the valid BIDS filenames into the ses-1/anat folder. The complete file structure to this point is pictured below.
###Organize files into folders
for files in $(ls sub*); do
Orgfile="${files%.*}"
Orgext="${files##*.}"
Modality=$(echo $Orgfile | rev | cut -d '_' -f1 | rev)
if [ $Modality == "T1w" ]; then
mv ${Orgfile}.${Orgext} ses-1/anat
else
:
fi
done
We have completed organizing the anat files. One can try confirming this is a validated BIDS dataset at this point. Once the validation is confirmed, we are ready to organize the diffusion files.
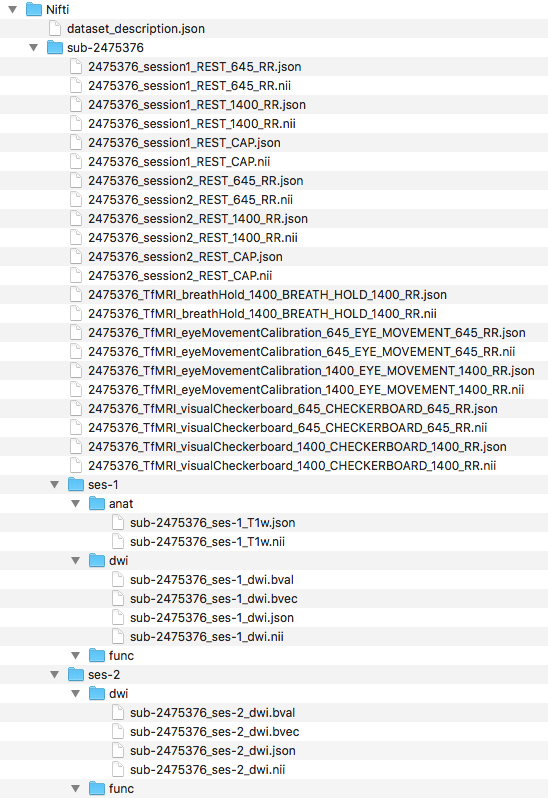
Step 5. To begin organizing the diffusion scans, we will generate the folder structure by creating a dwi folder within ses-1 and ses-2. Then we can convert the Dicom DTI folders within session1 and session2 and output the nii and json files to the participants Nifti folder. The output and current file structure can be seen below.
####Diffusion Organization####
#Create subject folder
mkdir -p ${niidir}/sub-${subj}/{ses-1,ses-2}/dwi
###Convert dcm to nii
#Converting the two diffusion Dicom directories
for direcs in session1 session2; do
${dcm2niidir}/dcm2niix -o ${niidir}/sub-${subj} -f ${subj}_${direcs}_%p ${dcmdir}/${subj}/${direcs}/DTI*
done
Next, we change the directory into the participants Nifti folder, where the converted nii and json files are.
#Changing directory into the subject folder
cd ${niidir}/sub-${subj}
Step 6. Now we can rename the diffusion nii and json files, similar to step 9 in part A. The original filename and BIDS filename are printed out.
#change dwi #Example filename: 2475376_session2_DIFF_137_AP_RR #BIDS filename: sub-2475376_ses-2_dwi #difffiles will capture how many filenames to change difffiles=$(ls -1 *DIFF* | wc -l) for ((i=1;i<=${difffiles};i++)); do Diff=$(ls *DIFF*) #This is to refresh the diff variable, same as the cases above. tempdiff=$(ls -1 $Diff | sed '1q;d') tempdiffext="${tempdiff##*.}" tempdifffile="${tempdiff%.*}" Sessionnum=$(echo $tempdifffile | cut -d '_' -f2) Difflast=$(echo "${Sessionnum: -1}") if [ $Difflast == 2 ]; then ses=2 else ses=1 fi mv ${tempdifffile}.${tempdiffext} sub-${subj}_ses-${ses}_dwi.${tempdiffext} echo "$tempdiff changed to sub-${subj}_ses-${ses}_dwi.${tempdiffext}" done
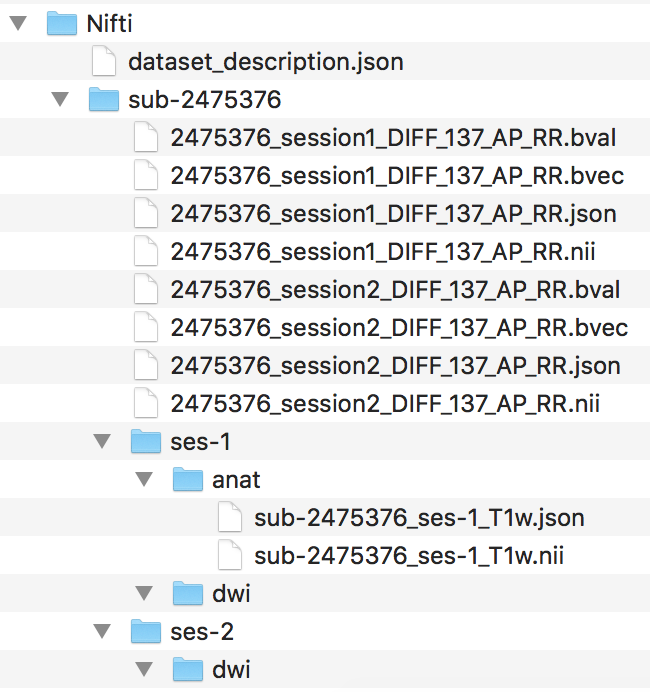
Step 7. After the filenames have been renamed, we can organize the files into their correct directories. The filenames and structure are visualized below.
###Organize files into folders
for files in $(ls sub*); do
Orgfile="${files%.*}"
Orgext="${files##*.}"
Modality=$(echo $Orgfile | rev | cut -d '_' -f1 | rev)
Sessionnum=$(echo $Orgfile | cut -d '_' -f2)
Difflast=$(echo "${Sessionnum: -1}")
if [[ $Modality == "dwi" && $Difflast == 2 ]]; then
mv ${Orgfile}.${Orgext} ses-2/dwi
else
if [[ $Modality == "dwi" && $Difflast == 1 ]]; then
mv ${Orgfile}.${Orgext} ses-1/dwi
fi
fi
done
We have completed the organization of the diffusion scans. One may confirm this is still a valid BIDS dataset through validation. Once validated, we are ready to organize the functional scans.
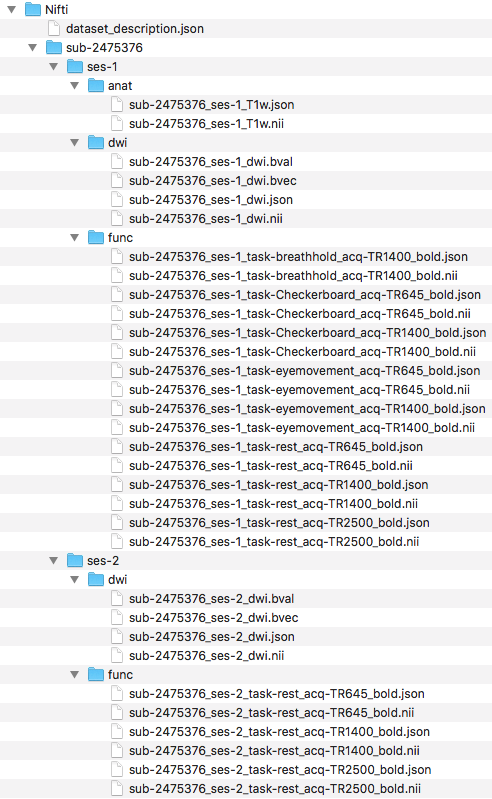
Step 8. To begin organizing the functional scans, we will create the functional folder structure by adding a func folder to both ses-1 and ses-2. With the folders created, we can now convert the functional dicom files to nifti and json files. To do this, we converted the folders that comprise the functional dicoms and output the nii and json files to within the participant’s folders in the Nifti directory. The folder names are contained within the for loop. The output and folder structure is visualized below.
####Functional Organization####
#Create subject folder
mkdir -p ${niidir}/sub-${subj}/{ses-1,ses-2}/func
###Convert dcm to nii
for direcs in TfMRI_breathHold_1400 TfMRI_eyeMovementCalibration_1400 TfMRI_eyeMovementCalibration_645 TfMRI_visualCheckerboard_1400 TfMRI_visualCheckerboard_645 session1 session2; do
if [[ $direcs == "session1" || $direcs == "session2" ]]; then
for rest in RfMRI_mx_645 RfMRI_mx_1400 RfMRI_std_2500; do
${dcm2niidir}/dcm2niix -o ${niidir}/sub-${subj} -f ${subj}_${direcs}_%p ${dcmdir}/${subj}/${direcs}/${rest}
done
else
${dcm2niidir}/dcm2niix -o ${niidir}/sub-${subj} -f ${subj}_${direcs}_%p ${dcmdir}/${subj}/${direcs}
fi
done
We changed the directory into where the converted nii and json files are, in the participant’s Nifti folder.
#Changing directory into the subject folder
cd ${niidir}/sub-${subj}
Step 9. Now we can rename the func files, similar to step 12 in part A. To do this, we changed filenames task by task. The order we renamed in: Checkerboard, eye movement, breath hold, and rest. Note that the rest scans still span across 2 sessions.
Checkerboard files renamed
##Rename func files #Break the func down into each task #Checkerboard task #Example filename: 2475376_TfMRI_visualCheckerboard_645_CHECKERBOARD_645_RR #BIDS filename: sub-2475376_ses-1_task-Checkerboard_acq-TR645_bold #Capture the number of checkerboard files to change checkerfiles=$(ls -1 *CHECKERBOARD* | wc -l) for ((i=1;i<=${checkerfiles};i++)); do Checker=$(ls *CHECKERBOARD*) #This is to refresh the Checker variable, same as the Anat case tempcheck=$(ls -1 $Checker | sed '1q;d') #Capture new file to change tempcheckext="${tempcheck##*.}" tempcheckfile="${tempcheck%.*}" TR=$(echo $tempcheck | cut -d '_' -f4) #f4 is the fourth field delineated by _ to capture the acquisition TR from the filename mv ${tempcheckfile}.${tempcheckext} sub-${subj}_ses-1_task-Checkerboard_acq-TR${TR}_bold.${tempcheckext} echo "${tempcheckfile}.${tempcheckext} changed to sub-${subj}_ses-1_task-Checkerboard_acq-TR${TR}_bold.${tempcheckext}" done
Eye movement calibration files renamed
#Eye Movement #Example filename: 2475376_TfMRI_eyeMovementCalibration_645_EYE_MOVEMENT_645_RR #BIDS filename: sub-2475376_ses-1_task-eyemovement_acq-TR645_bold #Capture the number of eyemovement files to change eyefiles=$(ls -1 *EYE* | wc -l) for ((i=1;i<=${eyefiles};i++)); do Eye=$(ls *EYE*) tempeye=$(ls -1 $Eye | sed '1q;d') tempeyeext="${tempeye##*.}" tempeyefile="${tempeye%.*}" TR=$(echo $tempeye | cut -d '_' -f4) #f4 is the fourth field delineated by _ to capture the acquisition TR from the filename mv ${tempeyefile}.${tempeyeext} sub-${subj}_ses-1_task-eyemovement_acq-TR${TR}_bold.${tempeyeext} echo "${tempeyefile}.${tempeyeext} changed to sub-${subj}_ses-1_task-eyemovement_acq-TR${TR}_bold.${tempeyeext}" done
Breath holding files renamed
#Breath Hold #Example filename: 2475376_TfMRI_breathHold_1400_BREATH_HOLD_1400_RR #BIDS filename: sub-2475376_ses-1_task-breathhold_acq-TR1400_bold #Capture the number of breath hold files to change breathfiles=$(ls -1 *BREATH* | wc -l) for ((i=1;i<=${breathfiles};i++)); do Breath=$(ls *BREATH*) tempbreath=$(ls -1 $Breath | sed '1q;d') tempbreathext="${tempbreath##*.}" tempbreathfile="${tempbreath%.*}" TR=$(echo $tempbreath | cut -d '_' -f4) #f4 is the fourth field delineated by _ to capture the acquisition TR from the filename mv ${tempbreathfile}.${tempbreathext} sub-${subj}_ses-1_task-breathhold_acq-TR${TR}_bold.${tempbreathext} echo "${tempbreathfile}.${tempbreathext} changed to sub-${subj}_ses-1_task-breathhold_acq-TR${TR}_bold.${tempbreathext}" done
Rest files renamed
#Rest
#Example filename: 2475376_session1_REST_645_RR
#BIDS filename: sub-2475376_ses-1_task-rest_acq-TR645_bold
#Breakdown rest scans into each TR
for TR in 645 1400 CAP; do
for corrun in $(ls *REST_${TR}*); do
corrunfile="${corrun%.*}"
corrunfileext="${corrun##*.}"
Sessionnum=$(echo $corrunfile | cut -d '_' -f2)
sesnum=$(echo "${Sessionnum: -1}")
if [ $sesnum == 2 ]; then
ses=2
else
ses=1
fi
if [ $TR == "CAP" ]; then
TR=2500
else
:
fi
mv ${corrunfile}.${corrunfileext} sub-${subj}_ses-${ses}_task-rest_acq-TR${TR}_bold.${corrunfileext}
echo "${corrun} changed to sub-${subj}_ses-${ses}_task-rest_acq-TR${TR}_bold.${corrunfileext}"
done
done
Next, we will organize the files into the correct directories. We have shown below, the filenames and organization.
###Organize files into folders
for files in $(ls sub*); do
Orgfile="${files%.*}"
Orgext="${files##*.}"
Modality=$(echo $Orgfile | rev | cut -d '_' -f1 | rev)
Sessionnum=$(echo $Orgfile | cut -d '_' -f2)
Difflast=$(echo "${Sessionnum: -1}")
if [[ $Modality == "bold" && $Difflast == 2 ]]; then
mv ${Orgfile}.${Orgext} ses-2/func
else
if [[ $Modality == "bold" && $Difflast == 1 ]]; then
mv ${Orgfile}.${Orgext} ses-1/func
fi
fi
done
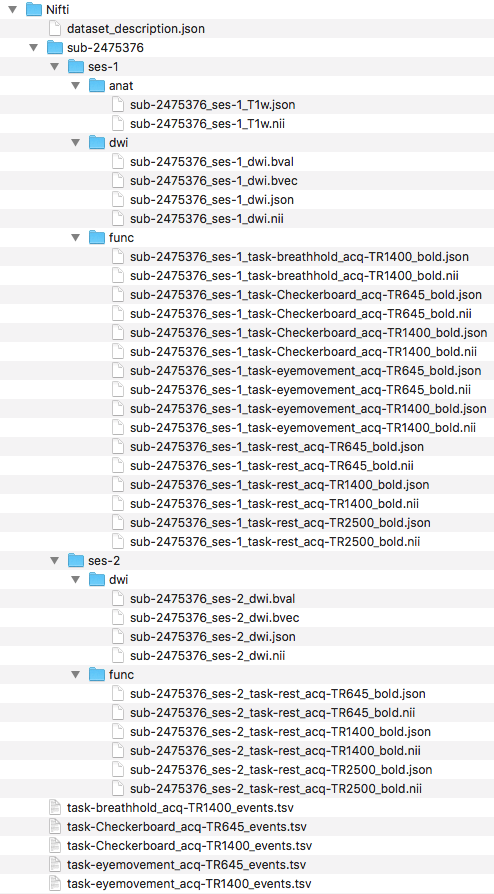
Step 10. Now we need to create the task event tsv files for each of our tasks. The task designs can be found on the NKI webpage. To determine the Checkerboard events file, one will look at the Checkerboard task design. Here we generated the Checkerboard TR=645 event file.
###Create events tsv files ##Create Checkerboard event file #Checkerboard acq-TR645 #Generate Checkerboard acq-TR645 event tsv if it doesn't exist if [ -e ${niidir}/task-Checkerboard_acq-TR645_events.tsv ]; then : else #Create events file with headers echo -e onset'\t'duration'\t'trial_type > ${niidir}/task-Checkerboard_acq-TR645_events.tsv #This file will be placed at the level where dataset_description file and subject folders are. #The reason for this file location is because the event design is consistent across subjects. #If the event design is consistent across subjects, we can put it at this level. This is because of the Inheritance principle. #Create onset column echo -e 0'\n'20'\n'40'\n'60'\n'80'\n'100 > ${niidir}/temponset.txt #Create duration column echo -e 20'\n'20'\n'20'\n'20'\n'20'\n'20 > ${niidir}/tempdur.txt #Create trial_type column echo -e Fixation'\n'Checkerboard'\n'Fixation'\n'Checkerboard'\n'Fixation'\n'Checkerboard > ${niidir}/temptrial.txt #Paste onset and duration into events file paste -d '\t' ${niidir}/temponset.txt ${niidir}/tempdur.txt ${niidir}/temptrial.txt >> ${niidir}/task-Checkerboard_acq-TR645_events.tsv #remove temp files rm ${niidir}/tempdur.txt ${niidir}/temponset.txt ${niidir}/temptrial.txt fi
Since both acquisitions have the same task design, we can simply copy the events file, but renamed.
##Checkerboard acq-TR1400 #Generate Checkerboard acq-TR1400 event tsv if it doesn't exist if [ -e ${niidir}/task-Checkerboard_acq-TR1400_events.tsv ]; then : else #Because the checkerboard design is consistent across the different TRs #We can copy the above event file and change the name cp ${niidir}/task-Checkerboard_acq-TR645_events.tsv ${niidir}/task-Checkerboard_acq-TR1400_events.tsv fi
Now, we will generate the event file for eye movement. We downloaded the eye movement paradigm. The path that we set for eye movement paradigm was: /Users/franklinfeingold/Desktop/EyemovementCalibParadigm.txt . This needs to be edited in the script to where one placed their paradigm file.
##Eye movement acq-TR645 #Generate eye movement acq-TR645 event tsv if it doesn't exist if [ -e ${niidir}/task-eyemovement_acq-TR645_events.tsv ]; then : else #Create events file with headers echo -e onset'\t'duration > ${niidir}/task-eyemovement_acq-TR645_events.tsv #Creating duration first to help generate the onset file #Create temponset file onlength=$(cat /Users/franklinfeingold/Desktop/EyemovementCalibParadigm.txt | wc -l) for ((i=2;i<=$((onlength-1));i++)); do ontime=$(cat /Users/franklinfeingold/Desktop/EyemovementCalibParadigm.txt | sed "${i}q;d" | cut -d ',' -f1) echo -e ${ontime} >> ${niidir}/temponset.txt done cp ${niidir}/temponset.txt ${niidir}/temponset2.txt echo 108 >> ${niidir}/temponset2.txt #Eye calibration length is 108 seconds #Generate tempdur file durlength=$(cat ${niidir}/temponset2.txt | wc -l) for ((i=1;i<=$((durlength-1));i++)); do durtime=$(cat ${niidir}/temponset2.txt | sed $((i+1))"q;d") onsettime=$(cat ${niidir}/temponset2.txt | sed "${i}q;d") newdur=$(echo "$durtime - $onsettime"|bc) echo "${newdur}" >> ${niidir}/tempdur.txt done #Paste onset and duration into events file paste -d '\t' ${niidir}/temponset.txt ${niidir}/tempdur.txt >> ${niidir}/task-eyemovement_acq-TR645_events.tsv #rm temp files rm ${niidir}/tempdur.txt ${niidir}/temponset.txt ${niidir}/temponset2.txt fi
Since the task design is consistent across different TR, we can simply copy the task design, but with a different filename.
##Eye movement acq-TR1400 #Generate eye movement acq-TR1400 event tsv if it doesn't exist if [ -e ${niidir}/task-eyemovement_acq-TR1400_events.tsv ]; then : else #Because the eye movement calibration is consistent across the different TRs #We can copy the above event file and change the name cp ${niidir}/task-eyemovement_acq-TR645_events.tsv ${niidir}/task-eyemovement_acq-TR1400_events.tsv fi
Lastly, we will create the event file for the breath hold task by looking at the breath hold design file.
##Breath hold acq-TR1400 #Generate breath hold acq-TR1400 event tsv if it doesn't exist if [ -e ${niidir}/task-breathhold_acq-TR1400_events.tsv ]; then : else #Create events file with headers echo -e onset'\t'duration > ${niidir}/task-breathhold_acq-TR1400_events.tsv #Create duration column #Creating duration first to help generate the onset file dur1=10 dur2=2 dur3=3 #Create tempdur file for ((i=1;i<=7;i++)); do echo -e ${dur1}'\n'${dur2}'\n'${dur2}'\n'${dur2}'\n'${dur2}'\n'${dur3}'\n'${dur3}'\n'${dur3}'\n'${dur3}'\n'${dur3}'\n'${dur3} >> ${niidir}/tempdur.txt done #Create onset column #Initialize temponset file echo -e 0 > ${niidir}/temponset.txt #Generate temponset file durlength=$(cat ${niidir}/tempdur.txt | wc -l) for ((i=1;i<=$((durlength-1));i++)); do durtime=$(cat ${niidir}/tempdur.txt | sed "${i}q;d") onsettime=$(cat ${niidir}/temponset.txt | sed "${i}q;d") newonset=$((durtime+onsettime)) echo ${newonset} >> ${niidir}/temponset.txt done #Paste onset and duration into events file paste -d '\t' ${niidir}/temponset.txt ${niidir}/tempdur.txt >> ${niidir}/task-breathhold_acq-TR1400_events.tsv #rm temp files rm ${niidir}/tempdur.txt ${niidir}/temponset.txt fi
Pictured below is the current file structure.
Step 11. At this point, one may try validating. However, one will receive the same error message from part A regarding defining TaskName in the task json files. The slice timing for multiband imaging was corrected in the version of dcm2niix implemented in the script.
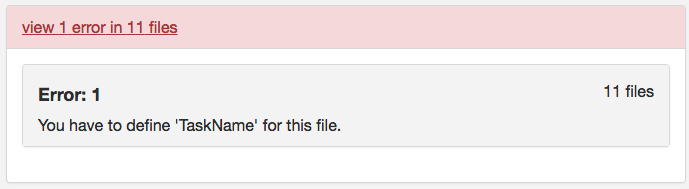
To correct this error, we will confirm that each task json file has the required fields: RepetitionTime, VolumeTiming or SliceTiming, and TaskName. This code snippet will evaluate if RepetitionTime exists, if SliceTiming (or VolumeTiming) exist and the timings are all less than the RepetitionTime, and if TaskName is defined. In addition, if TaskName is not defined, TaskName will be added to the json file with TaskName being the task label in the filename.
###Check func json for required fields #Required fields for func: 'RepetitionTime','VolumeTiming' or 'SliceTiming', and 'TaskName' #capture all jsons to test for sessnum in ses-1 ses-2; do cd ${niidir}/sub-${subj}/${sessnum}/func #Go into the func folder for funcjson in $(ls *.json); do #Repeition Time exist? repeatexist=$(cat ${funcjson} | jq '.RepetitionTime') if [[ ${repeatexist} == "null" ]]; then echo "${funcjson} doesn't have RepetitionTime defined" else echo "${funcjson} has RepetitionTime defined" fi #VolumeTiming or SliceTiming exist? #Constraint SliceTiming can't be great than TR volexist=$(cat ${funcjson} | jq '.VolumeTiming') sliceexist=$(cat ${funcjson} | jq '.SliceTiming') if [[ ${volexist} == "null" && ${sliceexist} == "null" ]]; then echo "${funcjson} doesn't have VolumeTiming or SliceTiming defined" else if [[ ${volexist} == "null" ]]; then echo "${funcjson} has SliceTiming defined" #Check SliceTiming is less than TR sliceTR=$(cat ${funcjson} | jq '.SliceTiming[] | select(.>="$repeatexist")') if [ -z ${sliceTR} ]; then echo "All SliceTiming is less than TR" #The slice timing was corrected in the newer dcm2niix version called through command line else echo "SliceTiming error" fi else echo "${funcjson} has VolumeTiming defined" fi fi #Does TaskName exist? taskexist=$(cat ${funcjson} | jq '.TaskName') if [ "$taskexist" == "null" ]; then jsonname="${funcjson%.*}" taskfield=$(echo $jsonname | cut -d '_' -f2 | cut -d '-' -f2) jq '. |= . + {"TaskName":"'${taskfield}'"}' ${funcjson} > tasknameadd.json rm ${funcjson} mv tasknameadd.json ${funcjson} echo "TaskName was added to ${jsonname} and matches the tasklabel in the filename" else Taskquotevalue=$(jq '.TaskName' ${funcjson}) Taskvalue=$(echo $Taskquotevalue | cut -d '"' -f2) jsonname="${funcjson%.*}" taskfield=$(echo $jsonname | cut -d '_' -f2 | cut -d '-' -f2) if [ $Taskvalue == $taskfield ]; then echo "TaskName is present and matches the tasklabel in the filename" else echo "TaskName and tasklabel do not match" fi fi done done
Step 12. Now if we try to validate, we find that this dataset is a valid BIDS dataset! To capture both subjects, one can change the subj for loop at the top of the script to replace participant 3893245 for 2475376 in the loop. After running participant 3893245 through the workflow, we find the same warnings as the part A curated dataset; the checkerboard scans across subjects have different time dimensions and the dwi may be missing scans because there is only 1 diffusion scan for participant 3893245 and 2 diffusion scans for participant 2475376. Note: Do not have both participants in the for loop if one has already been run and organized, this will cause an error.
This first tutorial part illustrated how to convert DICOMs from the NKI Test-Retest dataset to a validated BIDS dataset. We illustrated doing it A. manually and B. an automated custom solution. The next tutorial will show how to complete this conversation using an off-the-shelf BIDS converter: HeuDiconv.


Hi, Can you tell me if there is an amendment if you have MOCO files from a Siemens scanner?
Thank you for your question. To distinguish your MOCO file you may add to the filename the “rec” label. This rec label would be placed after task and acq (acq is optional). This can be found described here: http://bids.neuroimaging.io/bids_spec1.1.0.pdf#page=22
Hello, I am unable to create the dataset_description.json file using the jo command as it does not exist (I am a mac user). I can’t seem to find any other way to make my bash script do this, any suggestions?
Thank you for your question. To confirm, were you able to download and install the jo package successfully? The jo package can found here: https://github.com/jpmens/jo .
Vários modelos e cores de Tiaras e Faixas Para Bebês. https://blogprojeto.com/tiaras-para-bebe/
E ele vai voltar pq somos feitos de amor. http://shorl.com/brabibroprobefre
Hi, I have a question about pathway in this script:
####Defining pathways
toplvl=/Users/franklinfeingold/Desktop/NKI_script
dcmdir=/Users/franklinfeingold/Desktop/NKI_script/Dicom
dcm2niidir=/Users/franklinfeingold/Desktop/dcm2niix_3-Jan-2018_mac
#Create nifti directory
mkdir ${toplvl}/Nifti
niidir=${toplvl}/Nifti
####Anatomical Organization####
for subj in 2475376; do
echo “Processing subject $subj”
###Create structure
mkdir -p ${niidir}/sub-${subj}/ses-1/anat
###Convert dcm to nii
#Only convert the Dicom folder anat
for direcs in anat; do
${dcm2niidir}/dcm2niix -o ${niidir}/sub-${subj} -f ${subj}_%f_%p ${dcmdir}/${subj}/${direcs}
done
From my understanding dcmdir is a Dicom folder with sub folders for every subject. So I have a folder named Dicom on my computer with sub folders for each subject. Then a new folder on the Dicom level is created for new Nifti files. And subject 2475376 is a subfolder of Dicom with dicom files inside. With this set up the script is not running properly. The Nifty folder is being created with the structure but I am getting a message saying no dicoms are found for converting. Can you please explain how my folders should be set up? Where subjects should be placed etc.?
Hi Rachel,
Thank you for your question. To confirm, your dicom file structure should look like Dicom/2475376/anat/*.dcm . It may appear that the anat folder is missing.
The dicom file structure is shown in part 1A of this tutorial series: http://reproducibility.stanford.edu/bids-tutorial-series-part-1a/. For example, at step 5 I showed the file structure for the anatomical scan. The script is going into each scan sequence dicom directory. This is being done in the last section of the code you posted under the “#Only convert the Dicom folder anat” comment. The for loop is running through the anat folder to only convert the anatomical scan.
Hi Franklin,
I have no access to original dicom files, only nifti, .par, and .rec files. Is it possible to convert my files to BIDS format (automated way preferably) at this point? If it’s possible, what would be the way to go about this?
Thanks!
Paulina
Hi Paulina,
Thank you for your question! It is possible to convert your files to the BIDS format from the nifti files. (for the .par and .rec files they may be converted to nifti as well. I found this question that may explain how to do that – https://www.nitrc.org/forum/message.php?msg_id=11974). Once you have the nifti this tutorial shows how to rename and structure those niftis to follow the BIDS format. For example in anatomicals this would be following step 3 and 4. This may require writing a script that changes your current naming and structure to BIDS. Step 3 and 4 provide one way this may be done.
Thank you,
Franklin
I believe you are very conscious, funds allied with Germany
and except using the consent of Germany, we’re not able to seek peace with Spain without permission. So inside the summer, it
is possible to bring some sunscreen and insect repellents, sunshade hat and also other protection products;
but also in winter, you ought to wear some thick warm socks,
if required, you’ll be able to also bring a thermos bottle.
Such agencies will require complete responsibility of the print jobs and make you tension free.
This content is good, very interesting. Thank you for this great story.
Hey there, Im a software developer working in ISC888 Inc. in Thailand.
I have fond of web development, technology, and programming. I’m also interested in education and entrepreneurship.
Thanks for sharing a great article.
You are providing wonderful information, it is very useful to us.
Keep posting like this informative articles.
Thank you.
jokergaming-z
IF YOU NEED TO RICH CICK BELOW
slot-jokerz
Hello We’re jokergaming-z
if you need to rich click below
jokergaming-z
jokergaming-z
jokergaming-z
Welcome to new game pg auto slot
Hello We’re jokergaming-z
if you need to rich click below
jokergaming-z
jokergaming-z
jokergaming-z
It’s amazing Everything you write has meaning. I want to read in everyone.
Hello We’re jokergaming-z
if you need to rich click below
jokergaming-z
jokergaming-z
https://jokergaming-z.com
You’re write code pretty well.
I guess that you are great programmer.
I want to learning that from you.
This will be best experience for me.
Thanks for sharing this valuable content. It is very helpful for us.
Thanks for sharing, It is very helpful
Your post is very helpful to me. Thank you.
good information, it’s very helpful for me
I am interested in developing an TrafficMonitor GitHub application. This is very helpful, thank you very much.
Kasir Mandiri Aplikasi Kasir dan PPOB (Pulsa, Kuota, Voucher Game, PLN, Shopee Pay, Gopay, Dana)
arti kata, alquran indonesia, alquran english, asmaul husna, kamus bahasa inggris, kamus besar bahasa indonesia (kbbi), kamus bahasa gaul, antonim, sinonim, kamus kesehatan, kamus farmasi, kamus obat, kumpulan hadis, pdf editor (word to pdf, pdf to word, combine/merge pdf, split pdf, compress pdf, image/photo to pdf), kamus hukum, kamus bahasa jawa, kamus bahasa korea, kamus bahasa jepang, lirik lagu dan terjemahan bahasa indonesia, lirik lagu barat, resep masakan khas, resep masakan harian, resep masakan indonesia
got this site from my friend who shared with me regarding this website and
at the moment this time I am browsing this site and reading very
informative posts here
Nice article, thanks… https://finalank.org
Bookingtogo, online travel agen yang menyediakan tiket pesawat, hotel, tiket kereta api dan rekomendasi tempat wisata di Blog BookingToGo
joker slot has led the team to develop an online game system Let’s set up a team to develop slot into games that can be played online and can try playing slots. easily accessible through a connection in the internet.
Jackpots are easy to break, get money quickly, we pay for real
zeus 138 slot
Thank you for this very useful tutorial. I am new to BIDS and this tutorial really helped me to understand the basic concept of BIDS and how to use it.
Click on my name and you will have good luck
FineReader 2023. Sekarang ini ada banyak aplikasi perangkat lunak fantastis yang tersedia untuk dibeli. Mereka semua memiliki kemampuan untuk mengedit file PDF dan melakukan pengenalan karakter optik (OCR).Tetapi menurut kami, belum ada yang sehandal FineReader PDF.
Tidak ada perangkat lunak lain yang mungkin bisa setara dengan kombinasi hebat software ini. Mulai dari kemampuan mengerjakan dokumen PDF, OCR, maupun fitur perbandingan dokumen.Tidak ada aplikasi lain yang bisa mengintegrasikan semua komponen tersebut sejelas dan semulus aplikasi ini.
Pengguna perangkat ini dimanjakan untuk dapat mengkonversi, memodifikasi, dan mendistribusikan file PDF. Sangat cocok digunakan oleh individu maupun perusahaan. Teknologi ini berpotensi membantu tim di tempat kerja dalam merampingkan kolaborasi mereka dalam operasi PDF.
For Printer driver, you can visit: my-hpdrivers.com
Thanks alot and have good day
Withdraw money for real, fun and easy to win money.
qqslot777 login
Everyone’s Favorite Game
indo bet
It’s the same topic , but I was quite surprised to see the opinions I didn’t think of. My blog also has articles on these topics, so I look forward to your visit. luxury77
It’s the same topic , but I was quite surprised to see the opinions I didn’t think of. My blog also has articles on these topics, so I look forward to your visit. dewagg login
Withdraw money for real, fun and easy to win money.
idngg slot
A game that will make you earn money by playing on your mobile phone.
vegas88 asia
Everyone’s Favorite Game
elangslot
A very popular game camp that played many years ago. Until now because it’s a simple game. The rules are not complicated, the game is played for fun.
mantra88 slot
ทดลองเล่นยิงปลา Joker
TRY JOKER GAMING JOKER GAME We have a lot of games for you to choose from. If you’re going to play any game, you can play it all. Apply once.Play anytime, anywhere Download and install The application process is simple and hassle-free, as we value convenience. Speed and security for every candidate’s data.
Thankyou for taking the time to write this it was a great read. Good job!
slot5000 olympus
Absolutely outstanding information and very well written,thank you very much for this.
luxury 333
The game can go in unexpected directions at any time. It’s better to keep trying than nothing
gen77
Thank you for every other great post. The place else may just anyone get that type of info in such a perfect way of writing? I have a presentation next week, and I am at the look for such info.
Invite your friends to play this game
Play right away
angkasa138 slot
Play right away
megawin77
Invite your friends to play this game
prada88
More importantly, this game is open to play through the most suitable megagame website in 2023. Play สล็อต games by your own hand. How much you can play, pay just that. Plus many free bonuses. Press to receive by yourself. Don’t hold back. Deposit and withdraw through the automatic system. that makes a simple transaction in 3 seconds, that’s all.
Take your bonus now
Play and win now
bonus 168
Prepare yourself to win
agen388 login
Take your bonus now
hoki303 login
Let’s have fun, spin, enjoy, not boring with megagame, a stable website with an automatic system. Give away hard, give away for real There are many promotions, spinning, slot, easy, without interruption. There is a certificate from our government agency. Stable website confirms financial matters, definitely stable.
Many rewards await you
daftar judi cuan
Your luck awaits in our game
Try the game now
cuan 138 slot
This game is very exciting to play immediately
Let’s win this game
garuda99 login
When I have free time, I always come to study your articles. It’s really good. I want you to know.
hi!,I like your writing so much! proportion we https://www.zaargo.com.br/comprar-um-carro-em-uma-agencia-ou-a-um-particular/
One of the things I enjoy regarding reading websites such as this, is that there aren’t any spelling or lexical errors! Causes it to be tough about the readers sometimes. Very good work upon that and also the subject of this website. Many thanks!
Cryptocurrency Prices
Lemon: A Soothing Remedy for Sore Throat
A sore throat can be uncomfortable and bothersome, making it difficult to swallow or speak. Fortunately, there are natural remedies that can provide relief, and one such remedy is the humble lemon. Known for its tangy flavor and rich vitamin C content, lemons offer a range of benefits for soothing a sore throat. In this article, we will explore the reasons why lemon is good for a sore throat and how it can be used effectively to alleviate discomfort. So, let’s delve into the world of lemons and discover their remarkable properties.
Table of Contents
Introduction: Understanding the Power of Lemons for Sore Throats
Vitamin C: A Key Player in Soothing Sore Throats
Antibacterial and Antiviral Properties of Lemons
Soothing and Hydrating Properties of Lemon Juice
Natural Cough Suppressant
Honey and Lemon: A Dynamic Duo for Sore Throats
Lemon Tea: A Soothing Beverage for Sore Throats
Gargling with Lemon Water
Lemon Essential Oil: Inhalation for Relief
Precautions and Considerations
Conclusion
1. Introduction: Understanding the Power of Lemons for Sore Throats
Lemons, scientifically known as Citrus limon, are citrus fruits cherished for their refreshing taste and versatility. They are an excellent source of vitamin C, an essential nutrient known for its immune-boosting properties. When it comes to sore throats, lemons can provide relief and support the healing process.
2. Vitamin C: A Key Player in Soothing Sore Throats
Lemons are packed with vitamin C, which is known to strengthen the immune system and fight off infections. When you have a sore throat, your immune system is working hard to combat the underlying cause. Consuming lemons or lemon juice can provide a significant dose of vitamin C, helping your immune system function optimally and potentially speeding up the healing process.
3. Antibacterial and Antiviral Properties of Lemons
Lemons possess natural antibacterial and antiviral properties, which can be beneficial for a sore throat. The compounds found in lemons may help inhibit the growth of bacteria or viruses that could be causing the discomfort. By incorporating lemons into your remedies, you can provide your body with additional support in fighting off the infection and reducing inflammation.
4. Soothing and Hydrating Properties of Lemon Juice
One of the most noticeable benefits of using lemon for a sore throat is its soothing and hydrating properties. When mixed with warm water or tea, lemon juice can provide a soothing coating to the throat, alleviating dryness and irritation. Additionally, the hydration offered by lemon-infused drinks can help thin mucus and reduce discomfort.
5. Natural Cough Suppressant
Coughing is often associated with a sore throat and can further irritate the delicate tissues. Lemon’s natural cough-suppressant properties can help alleviate the urge to cough, providing temporary relief. Sipping on lemon-infused drinks can help soothe the throat and reduce coughing spells, allowing you to rest and recover more comfortably.
6. Honey and Lemon: A Dynamic Duo for Sore Throats
Combining lemon with honey creates a powerful remedy for a sore throat. Honey is known for its antibacterial and soothing properties, making it an excellent companion to lemon. Mixing warm water, lemon juice, and a teaspoon of honey creates a soothing elixir that can help reduce inflammation, coat the throat, and alleviate discomfort.
7. Lemon Tea: A Soothing Beverage for Sore Throats
Lemon tea is a classic remedy for sore throats, offering both the benefits of lemon and the soothing effects of warm liquids. To make lemon tea, simply squeeze fresh lemon juice into a cup of warm water and, if desired, add a teaspoon of honey for added sweetness. Sip on this comforting beverage throughout the day to soothe your throat and enjoy the benefits of vitamin C.
8. Gargling with Lemon Water
Gargling with lemon water can provide direct contact between the lemon’s beneficial compounds and the inflamed tissues in your throat. To make a lemon gargle solution, mix the juice of half a lemon with a cup of warm water. Gargle with this mixture for about 30 seconds before spitting it out. Repeat this process several times a day for relief from sore throat symptoms.
9. Lemon Essential Oil: Inhalation for Relief
Lemon essential oil, extracted from the lemon peel, can also be used to alleviate sore throat discomfort. Add a few drops of lemon essential oil to a bowl of hot water and inhale the steam. The soothing aroma of lemon can help reduce throat irritation and ease congestion. Ensure that you are using pure, high-quality essential oil for safe and effective use.
10. Precautions and Considerations
While lemons offer numerous benefits for sore throats, it’s essential to consider a few precautions. Lemon juice, when consumed in excess, can erode tooth enamel due to its acidic nature. To protect your teeth, it’s advisable to dilute lemon juice with water or consume it alongside a meal. Additionally, if you have any allergies or sensitivities to lemons, it’s best to avoid using lemon-based remedies.
11. Conclusion
Lemons are a natural and refreshing remedy for soothing sore throats. Packed with immune-boosting vitamin C, antibacterial properties, and a soothing effect, lemons can provide relief and support the healing process. Whether consumed as lemon water, lemon tea, or combined with honey, lemons offer a range of options to alleviate discomfort and promote a faster recovery. So, the next time you’re battling a sore throat, reach for a lemon and harness the power of this citrus gem.
lemon good for sore throat
I found your website very helpful. I especially liked the section above. See my profile
Get no code Adalo
I have recently started a blog, the info you offer on this site has helped me greatly. Thanks for all of your time & work.
Direct website channel, direct service that will answer every problem using labor with every platform.
So everyone would like to come and join in the entertainment with the production of income for the genuine direct website slots. Meet the needs of customers as needed. Forwarded for every day
เว็บพนันออนไลน์888เว็บตรง
Thanks for that article.
A really nice blog where we can found useful information.Thanks you.
Very informative article.
For Printer drivers, you can visit: https://www.sharpdrivers.net
Before you start the game, determine the amount of bet you want to place. Make sure you choose an amount that fits your budget and do not play beyond the specified limit.
perkasa777 slot
One of the best ways to increase your chances of winning slot games is to choose games that have high payouts. Don’t just choose games that look interesting but have less chance of winning.
login perkasa777
Klaycart is India’s best platform for dermatologist-recommended hair care & skin care products that help you to harmonize your external beauty with your inner radiance to make your world luminous, whole & complete. https://klaycart.com/products/irwings-spotlite
The HACCP system is a process control system guideline that applies to any organization those who are dealing with Manufacturing, trading, supply, retailing, packing, transportation, farming, etc of food products. It provides guidelines for identifying food safety Hazards, evaluating Food Safety Hazards, and Food safety Risk analysis. Get HACCP Certification with OSS Certification hassle-free and improve the business potential among your competitors.
https://www.osscertification.com/haccp-certification/
Thanks for your sharing, this very informative
Visit Sigarmas
I want to extend my sincere thanks for this helpful article. It’s been a lifesaver.
merhaba arkadaşlar, Makrome ipinden çok kolay örgü bileklik yapımı anlattım. kendini için veya hediye olarak sevdiklerinize…Mors Alfabesi Bileklik modelleri zarif ve şık görünümleri ile dikkat çekmektedir. Siz de evde kendiniz ya da sevdikleriniz için Mors Alfabesi Bileklik yapmak isterseniz yazımızda anlatacağımız kolay tekniği uygulayabilirsiniz. Bunun için tek yapmanız gereken zevkinize uygun renkte bir ip almak ve s düğüm Mors Alfabesi Bileklik yapımı için anlatacağımız Firuze Taşlı El Yapımı Makrome İpli Bileklik; Ayarlamalı olduğundan dolayı her bilek için uygundur. Çift Bileklikleri
I besides believe so , perfectly indited post! .
A game that will make you earn money by playing on your mobile phone.
https://virginiahighlandbb.com
Easy to trust site link indobet
When you lose something on a JetBlue plane, you can contact JetBlue Lost and Found department. To report a lost item on JetBlue flights, visit their official website and fill out an online lost item report with your contact information, flight details, and a description of the lost item. If there is no online form, contact JetBlue’s customer service directly. Remember to check with the airport’s Lost and Found and follow up with JetBlue periodically.
It’s the same topic , but I was quite surprised to see the opinions I didn’t think of. My blog also has articles on these topics, so I look forward to your visit. thesunshineskate.com
Unlock the best savings on all your favourite shops online! At Savzz, we’re committed to maximizing the value of your money. Whether you have a modest budget or a generous one, our extensive collection of discount codes, sales, and promo codes ensures that you can stretch your budget further. Utilizing our codes is a breeze – each one undergoes rigorous testing and verification to guarantee a seamless experience for you. Plus, rest assured that we consistently provide the latest discount codes in 2023, whether you’re shopping for a new wardrobe or upgrading your entertainment setup. Simply search for your preferred store and begin saving with confidence.
I besides believe so , perfectly indited post! . vegasslot77
Sparkling Success Starts Here: Unleash the power of Commercial cleaners in Melbourne. We make cleanliness your business advantage!
They shared their experiences as legal professionals, their https://bit.ly/3tAxv3H
Thank you for the information, don’t forget to visit my personal website at Afid Arifin.
This script Embossing Stamp is great for creating any text or graphics you want. It’s easy to use, just position the text or graphic and press down to emboss it.
We provide you with the best quality Cabinet Doors Refacing Toronto Building and Renovation Service you will ever experience. Our professional licensed teams are dedicated to providing you with a solution that satisfies your needs, meets your budget and exceeds your expectations. we provide exceptional customer service and will make sure to answer your questions and address any concerns you may have.
A Mortgage Loan Consultant in Perris, CA, serves as a crucial guide through the intricate landscape of home financing.
Agricultural Drones in Auburn Alabama have proven to be invaluable tools for farmers.
A CBD Vape Pen Canada is a convenient and discreet way to enjoy the benefits of cannabidiol (CBD). CBD is a non-psychoactive compound found in the hemp plant and has been shown to have numerous health benefits, including reducing anxiety, easing pain, and improving sleep.
A Jewish Wedding Music NYC brings joy, tradition, and celebration to the special day of a couple embarking on their journey of love.
Non Profit Organization in Brampton (NPO) is a business setup specifically to operate without any physical assets, liabilities or revenues for financial accounting purposes. It may be established with sound laws and bylaws but lacks credibility due to the lack of presence in market & proliferation of fake ones.
Celebrate a year of holistic fitness with Pilates Mat Exercises in Burnaby, BC. Nestled in the heart of this vibrant community, our studio has been a haven for wellness enthusiasts seeking a balance of strength and flexibility.
Fence Contractors Ottawa play a crucial role in transforming spaces and providing security, privacy, and aesthetic appeal to residential, commercial, and industrial properties.
Remote Notarization in USA has gained significant prominence, especially in a world shaped by digital transformations and the need for contactless solutions.
Basketball Academy Bar is the only exercise bar specifically designed for improving basketball skills and strength. Designed specifically for basketball players, players can develop all aspects of their game at home or in a gym with this superior performance training equipment. The smooth surface improves ball control while reducing slippage, while the non – skidding properties prevent falls. Players have access to courts on site 24/7, allowing them to practice whenever they want.
Gold Necklaces Women in Mississauga are a symbol of timeless elegance and refined taste, capturing the essence of sophistication and luxury for the modern woman. With their exquisite craftsmanship and exquisite designs, these necklaces serve as treasured heirlooms, reflecting the city’s diverse and dynamic fashion culture. Whether adorned with intricate filigree or sparkling gemstones, Gold Necklaces Women in Mississauga effortlessly enhance any ensemble, making a statement of both opulence and individual style, showcasing the fusion of tradition and contemporary flair that defines this vibrant Canadian city.
White Gold Earrings for Women in Chicago, Illinois epitomize a fusion of modern glamour and timeless allure, captivating those who seek refined extravagance in the heart of the Midwest.
From meticulous planning to diligent execution, this Office Moving Company in Yellowknife, NT, transforms the daunting task of relocation into a well-choreographed symphony, demonstrating their commitment to facilitating a stress-free transition for businesses in this vibrant northern community.
Bloor West Village offers a delightful escape into the world of pampering with its array of top-notch Manicure and Pedicure in Bloor West Village services.
Shawarma Calgary is an authentic Middle Eastern fast food restaurant offering a wide range of quality shawarma, kebab and donair sandwiches, platters and salads.
Doberman Puppies for sale in Tennessee offers an opportunity to welcome a devoted, loyal, and intelligent companion into your life.
Whether you’re looking to enhance your curb appeal or protect your investment from harsh weather conditions.Exterior House Painting Arizona has you covered.From prepping surfaces to selecting the perfect colors and paints.we take care of every detail to ensure superior results.
ensure superior results
Life is a game, so be a pro.
hoki368 login
This is a topic tһat iis nnear to my heart
zeus 138
time flies so fast, buddy
https://www.vkay.net/read-blog/48832_maximize-your-site-s-potential-with-high-quality-content.html
https://pt.linkedin.com/pulse/import%C3%A2ncia-das-plataformas-para-publicar-artigos-como-myrtys-maia-eykic?trk=public_profile_article_view
Thank you for the information, don’t forget to visit my personal website at link qq1221
In geometry, certain concepts are considered undefined terms as they do not have explicit definitions but are grasped through intuitive understanding. One such concept is the term “parallel lines.” To define parallel lines, a pair of specific undefined terms, “line” and “angle,” are employed. Parallel lines are defined as two lines in the same plane that never intersect, no matter how far they extend. This definition relies on the understanding of the undefined terms “line” and “angle.” By comprehending the fundamental properties of lines and angles, we can establish the concept of parallel lines and recognize their consistent distance apart. So, to answer the question, Which Pair of Undefined Terms is Used to Define the Term Parallel Lines? it is the combination of “line” and “angle” that forms the foundation of this geometric concept.
performance training equipment. The smooth surface improves ball control while reducing slippage, while the non – skidding properties prevent falls. Players have access to courts on site 24/7, allowing them to practice whenever they want.
Very impressive, this article explains the problem very clearly and completely. 100 from me and I really salute you! thank you, my respects:
Play and take big profits
slot5000 link
Hello” i can see that you are a really great blogger,
judi bola pandora188
Batın MR, manyetik alanlar ve radyo dalgaları kullanarak görüntüler elde etmek için bir cihaz kullanır. Batın MR, dokuların farklılıklarına dayalı olarak farklı sinyaller üretir ve bu sinyaller bilgisayar tarafından işlenerek üç boyutlu bir görüntü oluşturulur. Bu görüntüler, dokuların hassas bir şekilde görüntülenmesine izin verir ve dokuların sağlığı veya hastalığı hakkında ayrıntılı bilgi sağlar.
The popularity of online lottery as the best bet type and enthusiastic fans certainly can’t be denied anymore
luxury138
Wow, amazing post thank you. if you want more tech information click here.
indobet slot
nice article, i like ur site.
naga303 login
Invite your friends to win this game
login ligaciputra
very useful information
Rebahin
WaterApp’s Residential Water Level Sensor is installed into your tanks and wells and sends info to your phone via the cloud. It makes sure that you have enough safe water in your water tank. Receive alerts when the water level or quality is below the threshold or water gets overflowed or leaks. https://waterapp.in/technology/
Jackpots are easy to break, get money quickly, we pay for real
luxury333 slot
Thankyou for taking the time to write this it was a great read. Good job
Looking to secure reliable power solutions in Canada? Explore our range of 14kw Generac generators. Designed for Canadian conditions, these generators offer robust performance and uninterrupted power supply. Invest in peace of mind during outages with our 14kw Generac in Canada
good job for this information. situs gacor77
for me this is very helpful, thank you for sharing
Semir Sepatu
Incredibly insightful look forth to returning.
very nice post, i definitely enjoy this amazing site,
ingatbola88
Position yourself as a leader in Iraq with MaxiCert’s ISO Certification solutions. Build trust and credibility in your industry. Explore now: ISO Certification in Iraq.
Niche Information
rtp bonanza88 ➰ Daftar Website Slot Raja Bonanza88
Popularcert is your trusted partner in ISO certifications, dedicated to simplifying the certification process for businesses of all types and sizes. Our mission is to remove the complexity and confusion often associated with attaining ISO certification
ISO Certification in Saudi Arabia
Elevate your business standards in Saudi Arabia with PopularCert’s ISO Certification. Ensure quality and achieve success. Learn more: ISO Certification in Saudi Arabia.
you can see the amazing images like this in our blog birthday wishes images
A new and powerfull resource to follow latest technology news and articles. You can also find latest news and tutorials in my website.
A lifestyle magazine with losts of good and new posts. In this big website, we cover lots of interesting things. Check it!
“Insightful tutorial series! BIDS facilitates robust data management, crucial for achieving ISO certification standards in Oman, ensuring reproducibility and quality in research endeavors.”
ISO Certification in oman
This is an excellent piece with a wealth of information. Thanks for posting! also visit our website for NetSuite CRM services
I really got into this article. I found it to be interesting and loaded with unique points of interest. I like to read material that makes me think. Thank you for writing this great content.
This tutorial was a great introduction to the BIDS standard! The explanations were clear and the examples were helpful.
Customer satisfaction is a priority for Popularcert. We believe that a satisfied customer is the best testament to our success. Our dedicated customer service team is always ready to assist you with any queries or concerns, ensuring that your experience with us is positive and productive. We take pride in building long-lasting relationships with our clients, founded on trust and mutual respect.
https://popularcert.com/
Wow, this is fantastic content! It’s both informative and engaging. Thank you for sharing it.
This was very helpful. Thanks for sharing.
This article is fantastic! The information is incredibly useful, and I look forward to reading more from you.
Such valuable insights! I’m already looking forward to my next visit.
For businesses in Saudi Arabia aiming for ISO certification, partnering with Maxicert is a strategic move. Their comprehensive services pave the path for sustained success
Elevate your standards with Popularcert. Achieve ISO Certification in Saudi Arabia and access global markets.
Definitely a great effort! It’s encouraging to see such well-considered and articulate opinions written out. The writing was not only enjoyable, but also great. For more information and experienced support, please visit SEO Agency.