OpenNeuro App Highlights: ndmg-d
This post is part of a series of blog posts highlighting image analysis apps available on the OpenNeuro.org platform. It was contributed by Greg Kiar.
Welcome to part one of a two-part spotlight on an original BIDS App [1], Neurodata’s MRI to graphs (ndmg; pronounced nutmeg) pipeline for connectome estimation. The ndmg pipeline (made in Python, hosted on Github, packaged in Docker) has two distinct branches of connectome estimation: diffusion and functional. This part will highlight the diffusion branch currently available in OpenNeuro. Part two of this series will be published once the functional branch, ndmg-f, has been made publicly available through OpenNeuro as well.
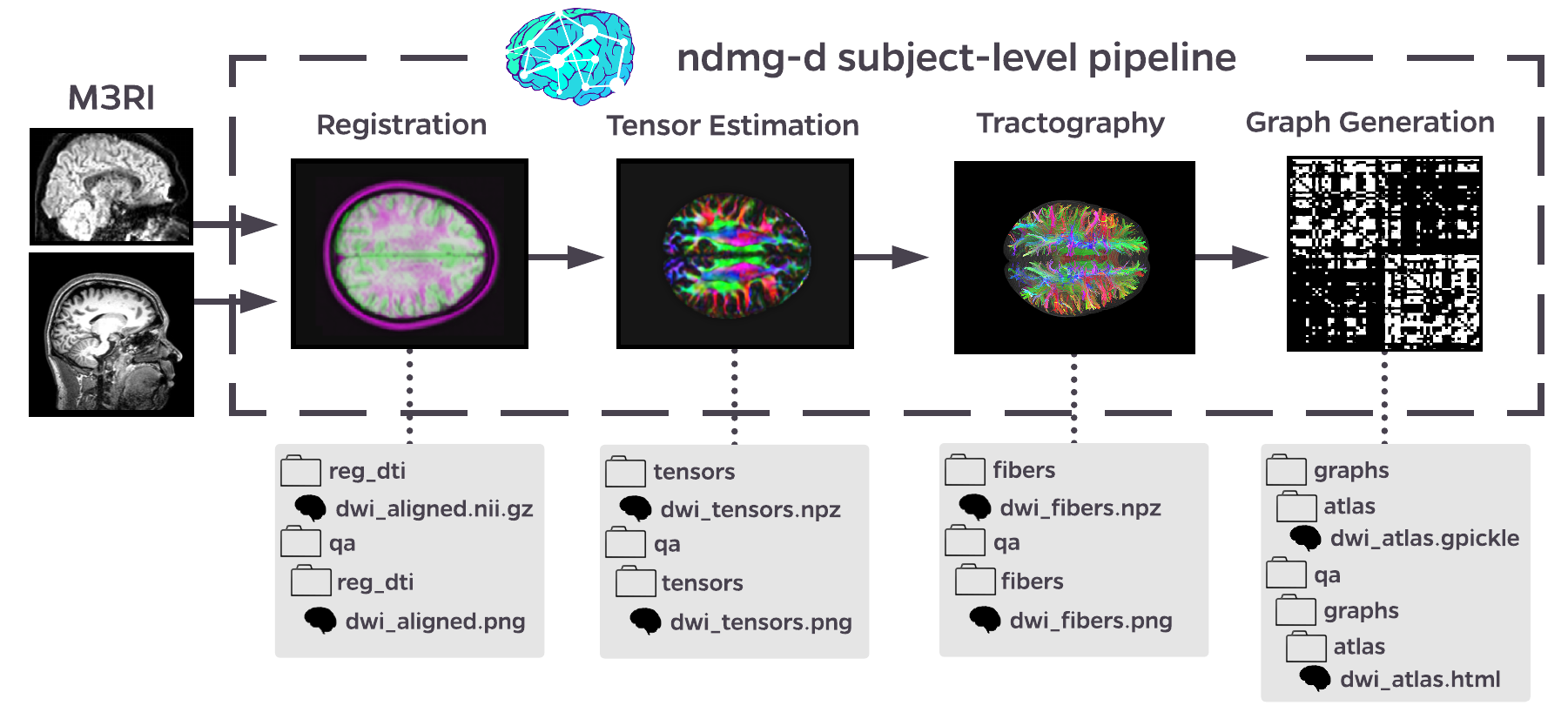
The ndmg pipeline is a one-click solution for reliable connectome estimation across a variety of scales [2]. Accepting as input BIDS datasets that include diffusion weighted MR images (DWI), corresponding parameter files (bval; bvec), and a structural scans (T1w, MPRAGE, etc.), the ndmg pipeline performs denoising, registration, tensor extraction, deterministic tractography, and ultimately graph generation across 24 parcellations in 1 mm MNI152 template space [3]. While graph generation code was developed in-house, denoising and registration leverage FSL [4], and tensor extraction and tractography leverage Dipy [5].
Figure 1: Schematic of ndmg
The ndmg pipeline takes approximately 1.5 hours to run, and has been tuned to operate robustly across a wide variety of diffusion imaging techniques, resolutions, and scanner types. The pipeline produces a rich array of quality control figures for intermediate steps in the pipeline and an interactive HTML plot the resulting connectomes in group-level analysis. These figures enable researchers to proof-read their results, spot outliers or potential failures easily, and serves as a springboard for exploratory analysis of the brain graphs.
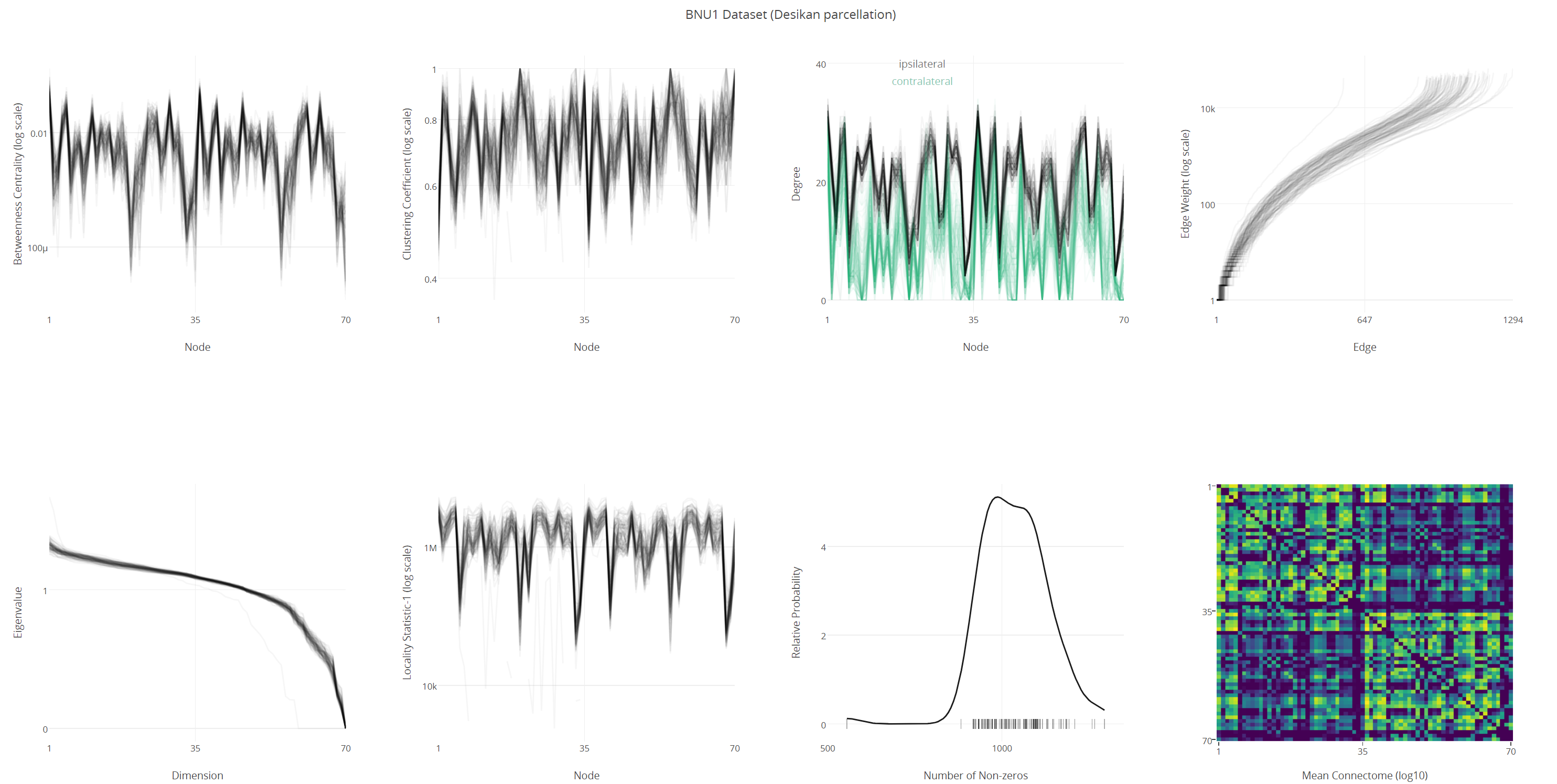
Figure 2: Graph summary statistics
The ndmg pipeline’s ultimate derivative is connectomes in the form of graph objects. Connectomes are generated by ndmg using 24 different brain atlases, ranging in size from ~50 to ~70,000 nodes. Intermediate derivatives may optionally be stored, using a debug flag, which include aligned DWI images, tensors, and fibers. An exemplar run on OpenNeuro can be found here.
The ndmg pipeline have been optimized over the discriminability of derived connectomes. In this context, discriminability is much like Test-Retest reliability: we report the probability that for any given session, the generated connectome is more alike a connectome generated from the same individual in another session than any other session from any other individual within the dataset. We computed this for several datasets, mainly within the CoRR [6] collection, with the peak and trough discriminability scores at 1.0 (perfect score), and 0.88, respectively, with most datasets scoring over 0.98.
We hope that this valuable integration with OpenNeuro and the BIDS specification increases the accessibility of the ndmg pipeline, and provides connectomics researchers with a reliable data processing solution in the cloud.
We encourage feedback, bug-reporting, and contributions on our Github repository! Happy processing!
References:
[1]: Gorgolewski, Krzysztof J., et al. PLoS computational biology 13.3 (2017): e1005209.
[2]: Kiar, Gregory, et al. bioRxiv (2017): 188706.
[3]: Fonov, Vladimir, et al. Neuroimage 54.1 (2011): 313-327.
[4]: Smith, Stephen M., et al. Neuroimage 23 (2004): S208-S219.
[5]: Garyfallidis, Eleftherios, et al. Frontiers in neuroinformatics 8 (2014).


This app is so useful