Big problems for common fMRI thresholding methods
A new preprint has been posted to the ArXiv that has very important implications and should be required reading for all fMRI researchers. Anders Eklund, Tom Nichols, and Hans Knutsson applied task fMRI analyses to a large number of resting fMRI datasets, in order to identify the empirical corrected “familywise” Type I error rates observed under the null hypothesis for both voxel-wise and cluster-wise inference. What they found is shocking: While voxel-wise error rates were valid, nearly all cluster-based parametric methods (except for FSL’s FLAME 1) have greatly inflated familywise Type I error rates. This inflation was worst for analyses using lower cluster-forming thresholds (e.g. p=0.01) compared to higher thresholds, but even with higher thresholds there was serious inflation. This should be a sobering wake-up call for fMRI researchers, as it suggests that the methods used in a large number of previous publications suffer from exceedingly high false positive rates (sometimes greater than 50%).
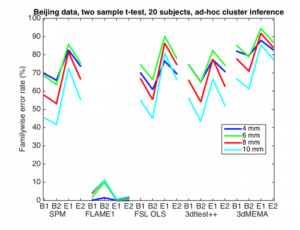
They also examined the commonly used heuristic correction (what they call “ad-hoc cluster inference”) of p=0.001 and a cluster extent threshold of 80 mm^3. This method showed a shockingly high rate of false positives, up to 90% familywise error in some cases. Hopefully this paper will serve as the deathknell for such heuristic corrections.
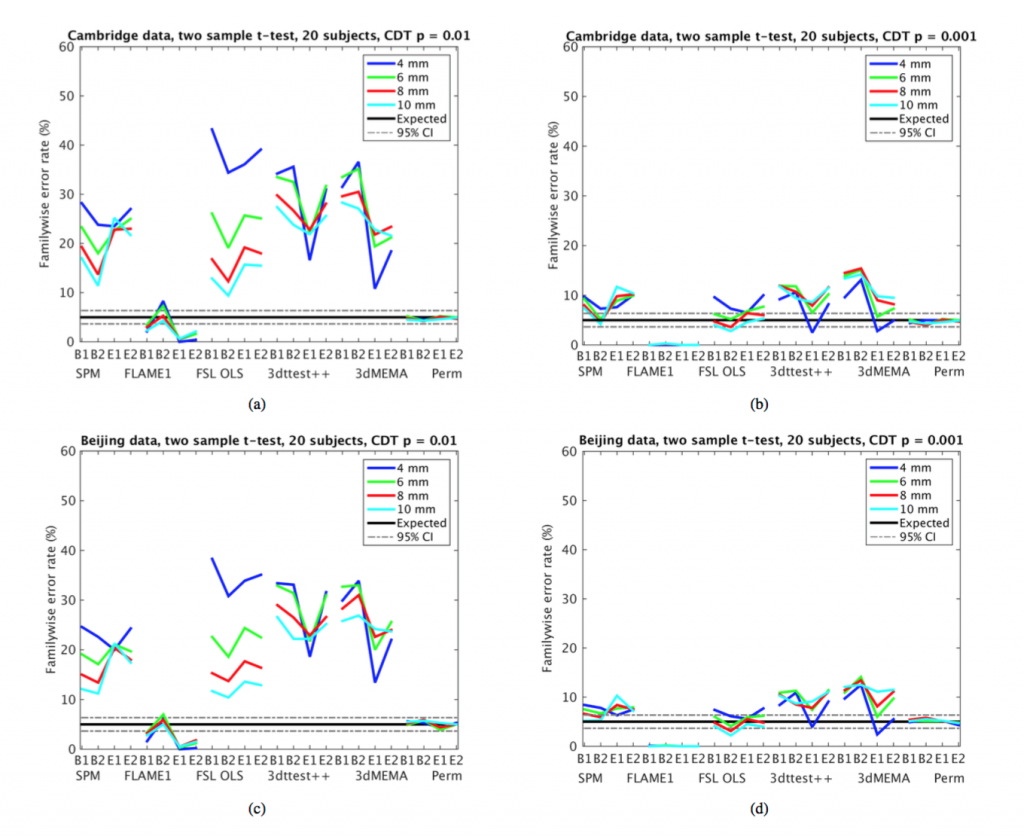
Another set of serious concerns were raised about the simulation-based methods implemented in AFNI’s 3DclustSim tool:
Firstly, AFNI estimates the spatial group smoothness differently compared to SPM and FSL. AFNI averages smoothness estimates from the first level analysis, whereas SPM and FSL estimate the group smoothness using the group residuals from the general linear model [39]. The group smoothness used by 3dClustSim may for this reason be too low (compared to SPM and FSL, see Figure 10); the variation of smoothness over subjects is not considered. Secondly, a 15 year old bug was found in 3dClustSim while testing the three software packages (the bug was fixed by the AFNI group as of May 2015, during preparation of this manuscript). The effect of the bug was an underestimation of how likely it is to find a cluster of a certain size (in other words, the p-values reported by 3dClustSim were too low).
I have been disturbed in recent years to see an increasing number of papers that perform most of their analyses using FSL or SPM, but then use the AFNI tool for multiple comparison correction. It is now clear why this tool was so attractive to so many researchers: It systematically undercorrects for multiple comparisons, leading to “better results” (i.e. inflated false positives). As an aside, this also points to the critical need for reporting of software versions in empirical papers; without this, it is impossible to know whether results obtained using AFNI suffered from this bug or not.
To the degree that there is a star of the Eklund et al. paper, it was nonparametric testing using permutation tests (for example, as implemented in the FSL randomise tool), though even those methods did not escape unscathed. The standard objection to nonparametric testing has been the substantial computational load of these methods. However, Eklund et al. also raise concerns about their ability to control Type I error rate, particularly in the one-sample t-test case (as opposed to the two-sample group comparison case where it did well). It appears that the assumption of symmetrically distributed errors required by the one-sample permutation approach may be violated, leading to inflated error, though these violations appear to be relatively minor compared to those seen with the parametric methods.
What are we to take away from this? First, the results clearly show that cluster-based inference with traditional parametric tools should always use cluster-forming thresholds no less stringent than p<0.001, as error inflation was much worse for less stringent cluster-forming thresholds. Second, the results strongly suggest that permutation testing is likely the best approach to prevent false positives without being overly stringent (e.g., using voxelwise inference rather that cluster inference). The platform being developed by the Center for Reproducible Neuroscience should make this much easier for researchers to apply through the use of high-performance computing. If one must use a parametric method, then FSL’s FLAME 1 with a cluster-forming threshold appears to be the best bet, though it is sometimes quite conservative. Third, the results should cause substantial suspicion about the use of AFNI’s 3DClustSim tool, as even the new version with the bug fix doesn’t bring false positive rates into line.
More generally, the fact that it took our field more than twenty years to discover that some of our most common methods are badly flawed is bracing. At the same time, it is only through the availability of massive open data repositories (including 1000 Functional Connectomes and OpenfMRI) that this kind of analysis could be done. We need much more work of the kind presented in the Eklund et al. paper in order to better understand how we can ultimately ensure that the results of fMRI studies are reproducible.


Hey come on. At least get Knutssons name right. It’s the least you could do.
Thanks for pointing this out – we have fixed the typo.