OpenNeuro App Highlights: MAGeT-Brain
This is a first episode of a series of blog posts highlighting image analysis apps available on the OpenNeuro.org platform. This piece was contributed by Gabriel A. Devenyi and Mallar Chakravarty.
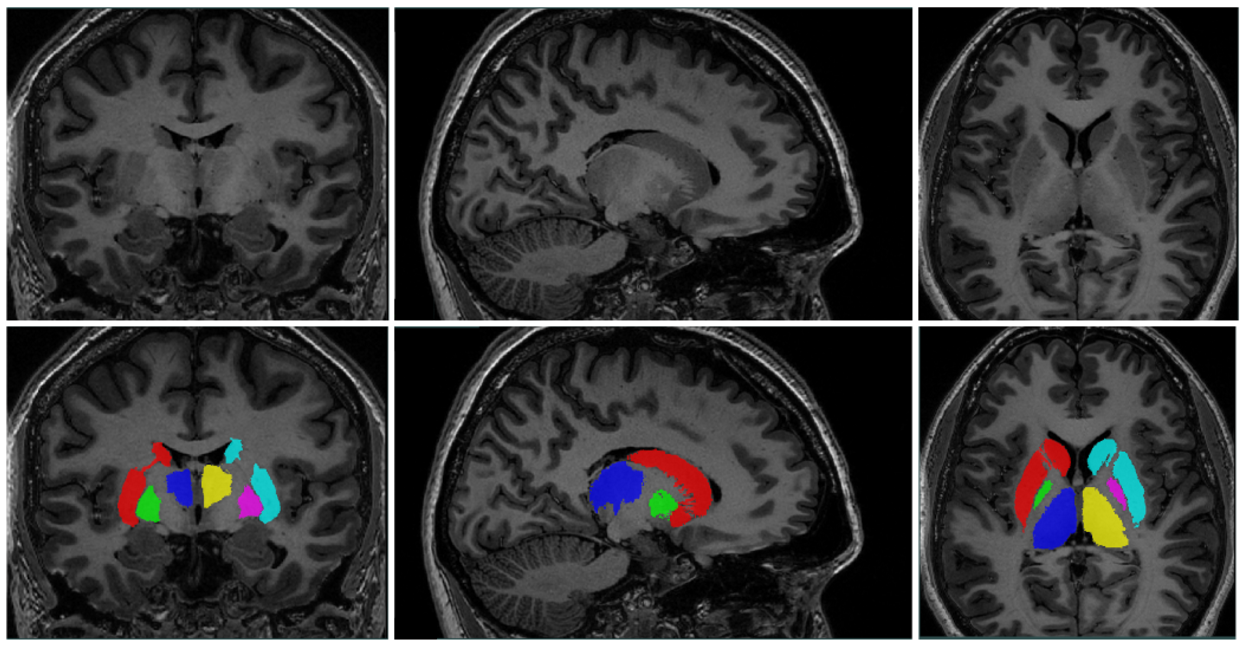
New to the OpenNeuro platform is the automatic structural segmentation pipeline MAGeT-Brain. Coming along with the pipeline are 5 expertly-segmented atlas/label combinations, at 0.3 mm isotropic, in T1 and T2, providing automatic segmentation of
- Subcortical structures: striatum, thalamus, globus pallidus
- Hippocampus subfields and hippocampal white matter
- Cerebellum and its lobules
- Amygdala
Figure 1: Example subcortical segmentation
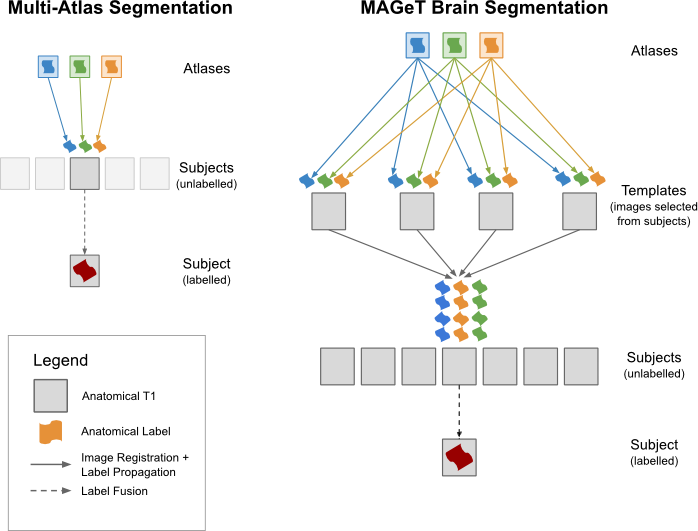
MAGeT-Brain is an extension of the classic registration based “multi-atlas” segmentation method, where labels on manually segmented atlases are transformed via linear/non-linear registration onto a set of subjects. The resulting candidate segmentations are combined via some label fusion technique.
Figure 2: Classical multi-atlas vs. MAGeT-Brain segmentation
The innovation of MAGeT-Brain lies in the introduction of the “template” layer, a representative subset of the subject population, which is used to increase the anatomical variance in the atlas set. These new template brains are used to produce the candidate segmentations on the subject brains, resulting in a much larger total number of candidates and overall better segmentations. Such a pipeline is relatively computationally expensive, requiring cluster computing to produce results in a reasonable amount of time. MAGeT-Brain has been validated and compared to other leading segmentation tools and found to be superior for low atlas counts, and equivalent for higher atlas counts. Since the production of high-resolution, anatomically-informed manual segmentations is very labour intensive, MAGeT-Brain’s computational load is a reasonable tradeoff to produce optimal segmentations.
Previously MAGeT-Brain was implemented to support the specialized MINC2 file format. However, this pipeline was recently rewritten to support all file formats supported by ITK and ANTs, allowing integration into BIDS-Apps and OpenNeuro via a small runtime shim.
We welcome our new users of OpenNeuro and hope you find this tool useful!
References:
Treadway MT, Waskom ML, Dillon DG, et al. Illness Progression, Recent Stress, and Morphometry of Hippocampal Subfields and Medial Prefrontal Cortex in Major Depression. Biol Psychiatry. 2014;
Entis JJ, Doerga P, Barrett LF, Dickerson BC. A reliable protocol for the manual segmentation of the human amygdala and its subregions using ultra-high resolution MRI. Neuroimage. 2012;60(2):1226-35.
Park, M.T., Pipitone, J., Baer, L., Winterburn, J.L., Shah, Y., Chavez, S., Schira, M.M., Lobaugh, N.J., Lerch, J.P., Voineskos, A.N., Chakravarty, M.M. High-resolution atlases of the cerebellum and cerebellar lobules at 3T: Derivation and automated segmentation using multiple automatically generated templates. NeuroImage (2014)
Chakravarty MM, Bertrand G, Hodge CP, Sadikot AF, Collins DL. The creation of a brain atlas for image guided neurosurgery using serial histological data. Neuroimage. 2006;30(2):359-76.
Winterburn JL, Pruessner JC, Chavez S, et al. A novel in vivo atlas of human hippocampal subfields using high-resolution 3 T magnetic resonance imaging. Neuroimage. 2013;74:254-65.
Amaral, R.S.C., Park, M.T.M., Devenyi, G.A., Pruessner, J.C., Pipitone, J., Winterburn, J., Chavez, S., Schira, M., Lobaugh, N., Voineskos, A.N., Chakravarty, M.M, and Alzheimer’s Disease Neuroimaging Initiative (2016). Manual segmentation of the fornix, fimbria, and alveus on high-resolution 3T MRI: Application via fully-automated mapping of the human memory circuit white and grey matter in healthy and pathological aging. NeuroImage.
Pipitone J, Park MT, Winterburn J, et al. Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014;
M Mallar Chakravarty, Patrick Steadman, Matthijs C van Eede, Rebecca D Calcott, Victoria Gu, Philip Shaw, Armin Raznahan, D Louis Collins, and Jason P Lerch. Performing label-fusion-based segmentation using multiple automatically generated templates. Hum Brain Mapp, 34(10):2635–54, October 2013.


Submit a comment