fMRIPrep tutorial: Running the docker image
Introduction
Welcome to our tutorial on running fMRIPrep (for a more detailed description of the tool, please check out our recent paper/preprint). In this tutorial, we will be utilizing the docker container of fMRIPrep. This tutorial will illustrate a detailed step-by-step guide on how to prepare and run this. We will provide the command line inputs to do this. The guide will be using data prepared and BIDS validated from a previous tutorial.
Table of contents
- Setting up your environment
- FreeSurfer License
- Running fMRIPrep
- fMRIPrep outputs and the visual reports
- Troubleshooting
Guide to running fMRIPrep
Step 1. To begin, we first need to initialize the environment. We will be utilizing Docker for running fMRIPrep. In addition, please download Python. After you have gotten Docker and Python, we can use the command line to download a wrapper to run fMRIPrep. The command is shown below.
pip install --user --upgrade fmriprep-docker
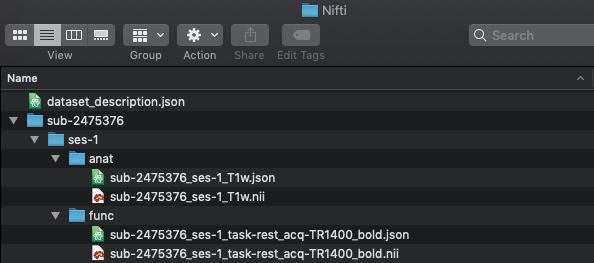
For this tutorial, we will be using the Nathan Kline Institute (NKI) Rockland Sample – Multiband Imaging Test-Retest Pilot Dataset. We will only be taking sub-2475376 session 1 anatomical and resting state scan at TR=1400ms. Please find pictured below our BIDS validated dataset.
NOTE: Please observe there are a few BIDS validator warnings that may need to be attended to before running fMRIPrep.
1) To do the susceptibility distortion correction, the IntendedFor metadata keys need to be defined for every field map that is to be applied.
2) To do the slice timing correction, the slice timings need to be defined in the BOLD json metadata file (if you want to ignore the slice timing correction, please add the ‐‐ignore slicetiming option, the command to do this is shown below.)
3) Please be cautious running multiple subjects in parallel due to space and memory limits on your machine.
Step 2. fMRIPrep uses FreeSurfer to reconstruct anatomical surfaces of the brain as well as some registration steps. Because of this, fMRIPrep requires a FreeSurfer license. To get the license and run FreeSurfer, FreeSurfer requires users to register for free, here. After filling out the registration form, a license file will be sent to the email provided. Please save this license file to your computer. We saved our license into a file called /Users/franklinfeingold/freesurfer.txt. More information regarding this can be found here.
Step 3. Now we can call fmriprep-docker. The command to execute it is illustrated below. To break down the command: the first argument is providing the input path to the BIDS dataset. The next part is providing the output directory. Followed by passing the keyword ‘participant’ as the third argument, which selects the analysis level that will be executed (see the BIDS-Apps specification for further details and possible values; in the case of fMRIPrep, only ‘participant’ is available). Lastly, we provide the participant label tag to indicate which participant to run. Please note that the ‘sub-’ prefix is not included. You may pass additional arguments as you would to fMRIPrep. For simplicity, we recommend placing the input, output and analysis levels prior to any options.
fmriprep-docker /Users/franklinfeingold/Desktop/BIDS_NKI_walkthrough/Nifti /Users/franklinfeingold/Desktop/BIDS_NKI_walkthrough/derivatives participant --participant-label 2475376 --fs-license-file /Users/franklinfeingold/freesurfer.txt
Running this command may download the latest docker image of fMRIPrep if you don’t have it installed already. This installation will take time and the screen may not update often, but the process is running. We are also checking for memory constraints and you may need to increase your docker memory. In the command line it will output a “RUNNING: “ line. This is showing the docker command that is actually constructed and ran. If the fmriprep-docker “command not found” this response will be helpful to solve this issue.
fMRIPrep is now running on your data!
If you would like to add additional options to the call, this is the step to do this. For example, if you would like to ignore the slice timing correction. The call is shown below.
fmriprep-docker /Users/franklinfeingold/Desktop/BIDS_NKI_walkthrough/Nifti /Users/franklinfeingold/Desktop/BIDS_NKI_walkthrough/derivatives participant --participant-label 2475376 --fs-license-file /Users/franklinfeingold/freesurfer.txt --ignore slicetiming
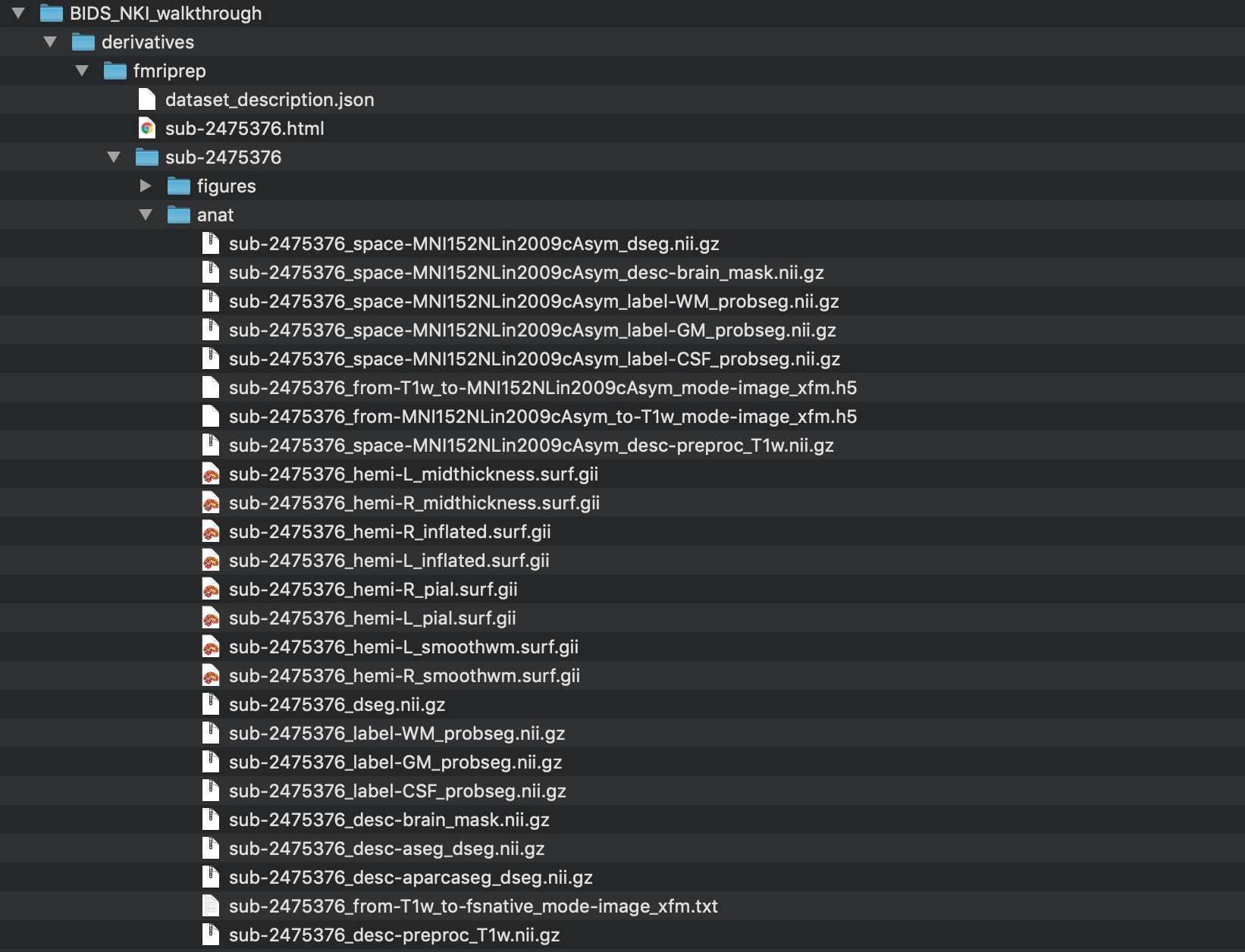
Step 4. When fMRIPrep has completed running, the output directory looks like this.
A detailed description of the fMRIPrep outputs can be found here. More broadly, the outputs of fMRIPrep conform to the BIDS-Derivatives specification, Release Candidate 1.
NOTE: fMRIPrep does not run confounds regression or any other filtering. We do provide a confounds .tsv file with one confounding time-series per column, from which you can select and use after pertinent orthogonalizations to regress confounds out with their preferred tools.
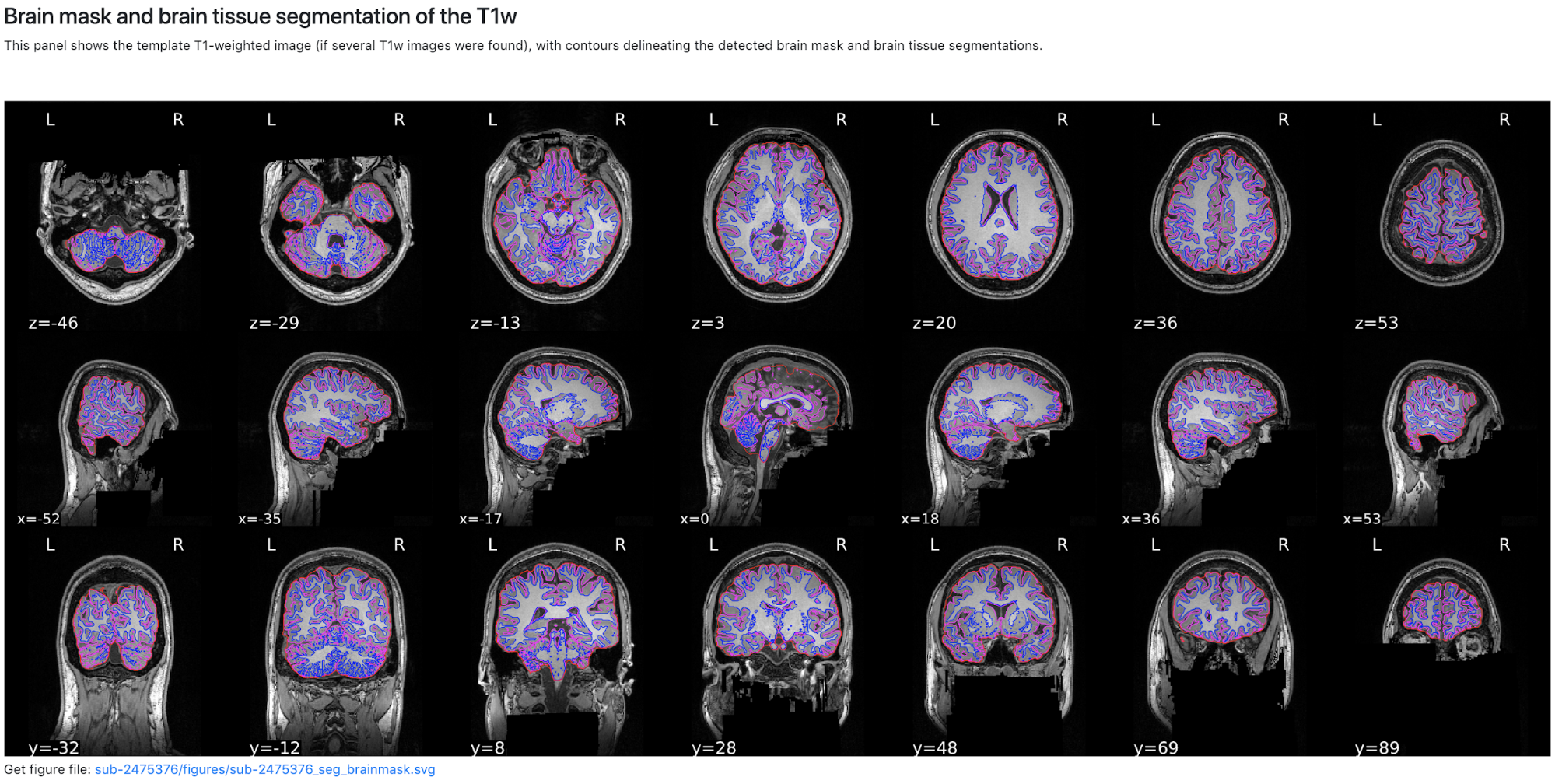
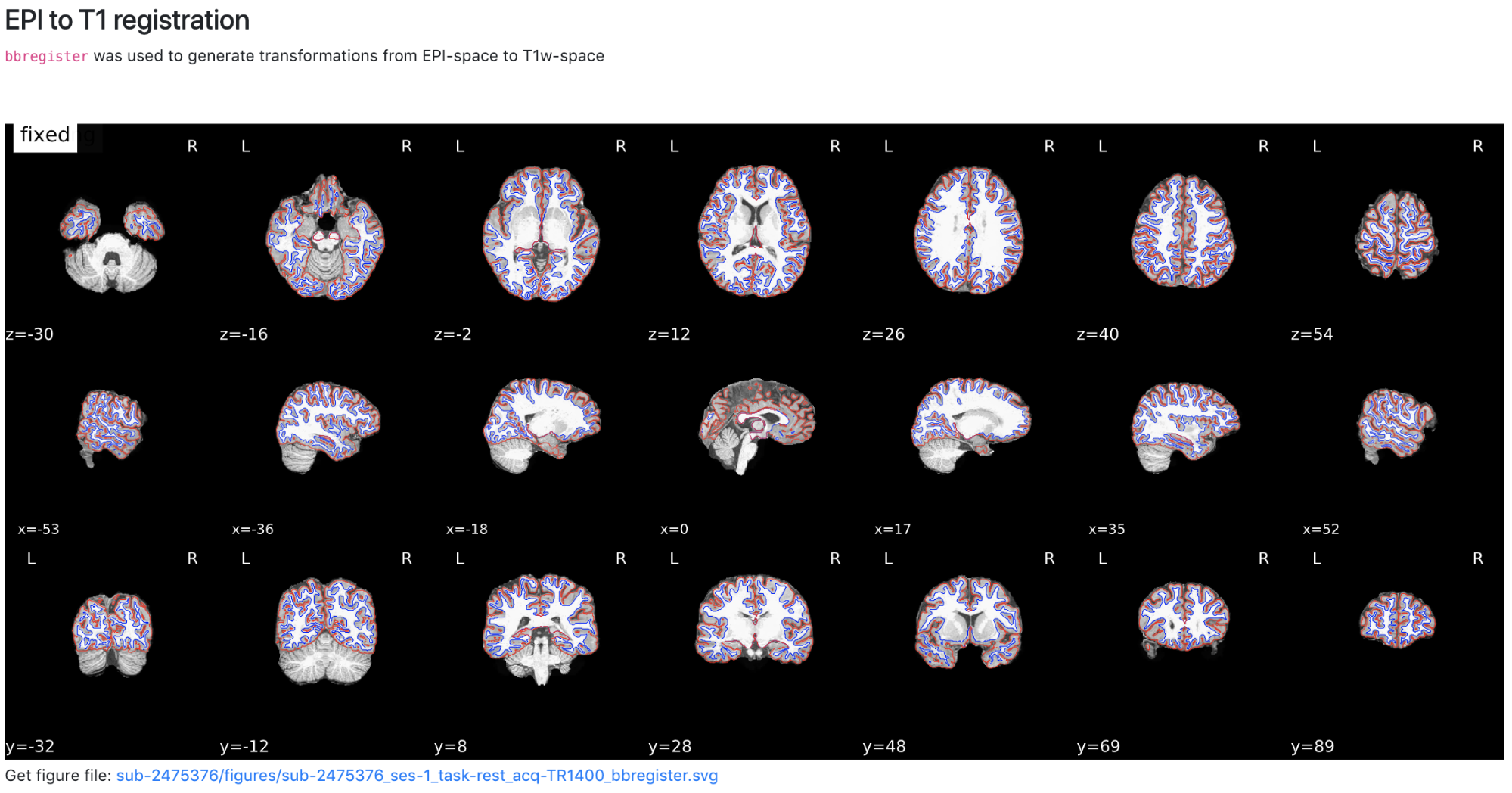
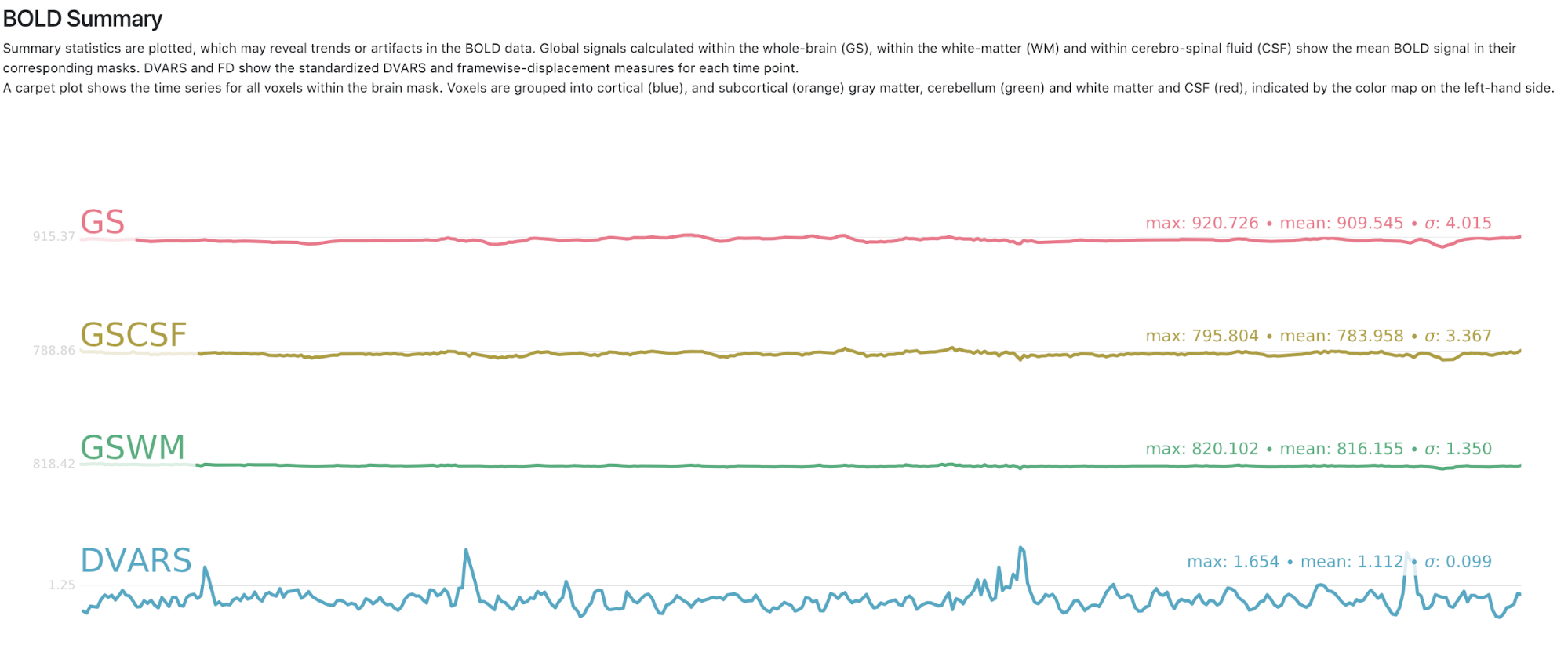
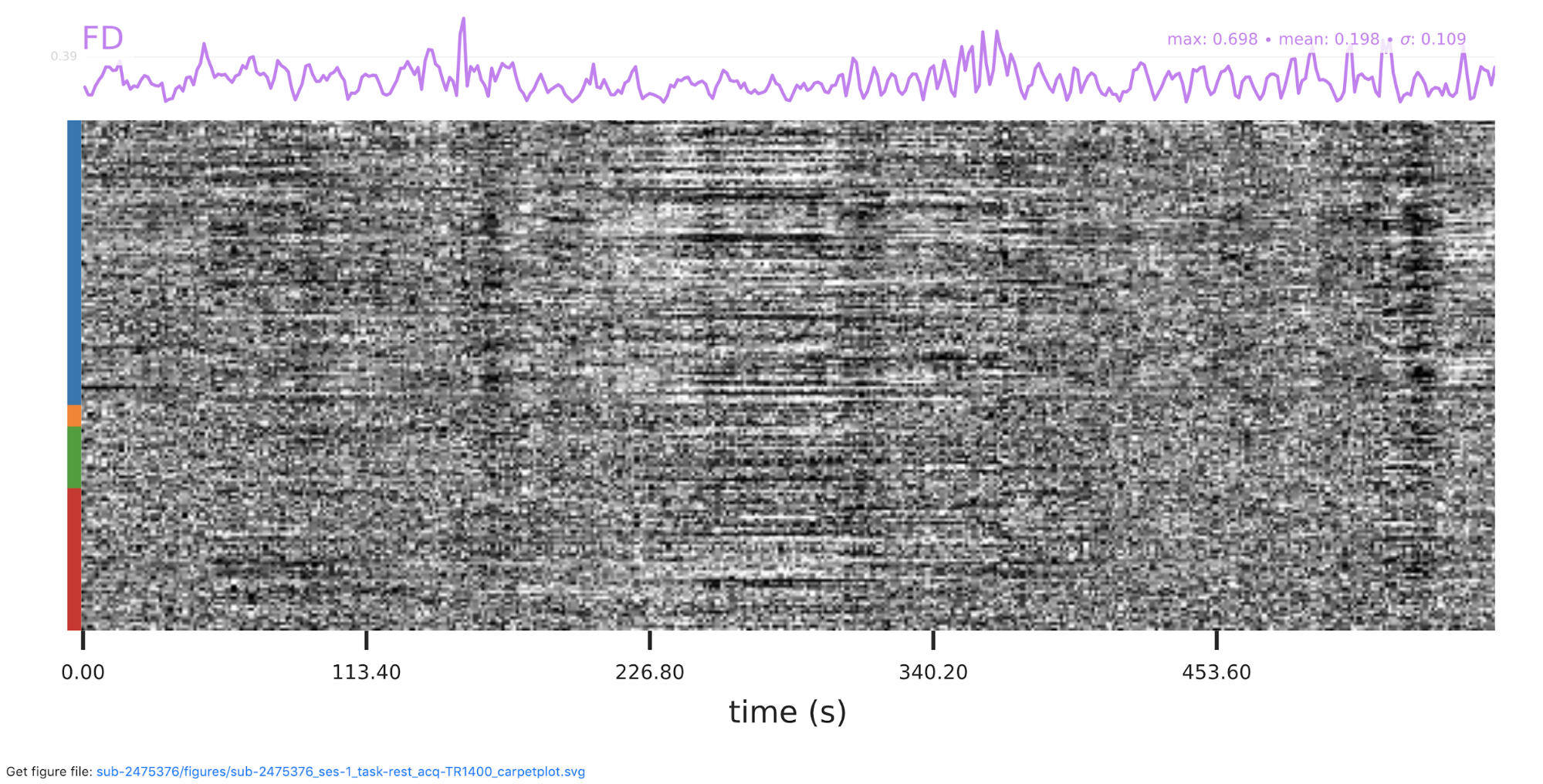
Now we have completed running fMRIPrep on the dataset! What’s Next? After running fMRIPrep we encourage you to open up the generated visual reports (for example, screenshots of the Brain mask and brain tissue segmentation of the T1w, EPI to T1 registration, and the BOLD Summary portions are shown below). These reports, written in HTML to be viewed in your web browser (we recommend Chrome or Firefox), will walk you through the preprocessing steps run, and highlights some of the decisions fMRIPrep made along the way. A careful examination of the report of every subject should be done to assess the quality of the preprocessing results. Under the “Methods” section of each report, you’ll find a detailed description of the exact processing workflow ready to copy-and-paste into your manuscript, including all pertinent references.
Brain mask and brain tissue segmentation of the T1w
Step 5. New users may run into two families of problems:
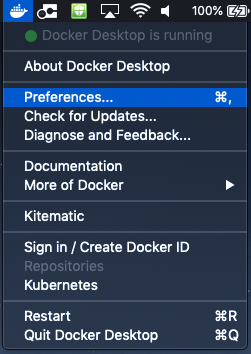
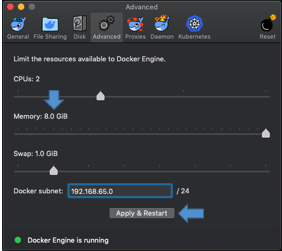
- Memory issues: on Windows and Mac OSX systems, the Docker Engine is configured to access 2GB RAM by default. Please, check your Docker configuration before running fMRIPrep. We recommend assigning 8GB or more memory to the Docker Engine. Please make sure to apply & restart or this change will not have been implemented. To check/change this we have shown the process below.
- Mounting drives on Windows: the Docker for Windows Engine does not allow sharing data from the host to make it available within the container. fMRIPrep requires that the original data are “mounted” into the container. Please make sure you allow Docker to mount folders from the host in your settings.
- fMRIPrep requests the FreeSurfer license file, although one is provided. If you are on Windows, you may experience this problem for the same reason immediately above. Please check your drive-sharing permissions.
This tutorial demonstrated how to run fMRIPrep on a NKI subject. If you have any questions, please direct them to neurostars using the fmriprep tag and the experts will be able to help you through the issue. If you would like to contribute to fMRIPrep, please find the GitHub repository here.


How does this work on windows?
Hi,
Thank you for your question. The installation and configuration may be the most challenging. On the fMRIPrep documentation, there is a mention of what to be beware of: https://fmriprep.readthedocs.io/en/stable/installation.html?highlight=windows. As you are running through this, if you have any questions please ask on NeuroStars: https://neurostars.org/tags/fmriprep. The experts will be able to answer your specific questions through the process.
Thank you,
Franklin
Thank you for this tutorial. I want to know whether there are tutorials about how to interpret the visual reports. I have used fMRIPrep to do the preprocess, but I am not sure whether the results are fine.
Thank you for your time.
Zhenfu
Hi Zhenfu,
Thank you for your question and happy to hear your use of fMRIPrep. There are likely to be recommendations/advice on https://neurostars.org/tags/fmriprep (this is a forum where you may ask questions and read issues other users have flagged). Currently, I am not aware of specific tutorials to interpret the reports.
Thank you,
Franklin
Is there a BIDS formatted version of the tutorial data? I would rather not go through the whole BIDS tutorial just to have a reference dataset as I work on getting fMRI prep and podman properly configured on RHEL.
Hi Chris,
Thank you for your question. There is not a BIDS formatted version of the tutorial data available. The BIDS tutorial 1B – http://reproducibility.stanford.edu/bids-tutorial-series-part-1b/ – provides the commands to convert and streamline the process. This tutorial only uses a smaller sample of that tutorial to make it easier to get to this stage. Another option is to grab data off of https://openneuro.org/. Datasets need to be BIDS validated, but depending on what options you may be looking to see (i.e. Slice Timing correction, the timing would need to be defined in the json to do the correction). I apologize for the inconvenience.
Hi,
Thank you for the tutorial. It helped me how to use fmriprep.
Now I need to process Multi-echo data using fMRIprep. Do you have similar type of fMRIPrep tutorial for MUlti-echo data.?
Thank you
Sameera
Hi Sameera,
Thank you for your question! There is not a specific tutorial for Multi-echo data, perhaps on NeuroStars with tag fmriprep (https://neurostars.org/tags/fmriprep), there are useful comments from the community. Please feel free to post your question on NeuroStars, fmriprep experts are on there and able to help!
Thank you,
Franklin
Hi,
how much does it take usually for the process to be done completely on a single subject? On my local I have 8 cores and 16 GB of RAM. It has been running for more than 5 hours now and it’s not finished yet. Considering I am running fMRIPrep on just one subject!
And another question I’m asking is how may I modify fMRIPrep for Macaque monkeys brain? I didn’t find any description about this in the prepared docs!
Hi Matin,
Thank you for your messages. I would suggest asking these over on NeuroStars using the fMRIPrep tag (https://neurostars.org/tags/fmriprep). fMRIPrep experts are on NeuroStars to help answer questions.
Thank you,
Franklin
Thanks for this helpful tutorial. Can you think of any reason why after running command in Step 3, no directory called “fmriprep” was created? I still get the “sub-/” directory and others but somehow the structure is different.
Hi,
Thank you for the tutorial. It helped a lot.
I want to konw how to execute with Docker in parallel.
Thank you for your time.
Wenjun
Hello We’re jokergaming-z
if you need to rich click below
jokergaming-z
jokergaming-z
jokergaming-z
Nice article, thanks.. https://finalank.org